NPs Basic Information
|
Name |
Cyclobarbital
|
| Molecular Formula | C12H16N2O3 | |
| IUPAC Name* |
5-(cyclohexen-1-yl)-5-ethyl-1,3-diazinane-2,4,6-trione
|
|
| SMILES |
CCC1(C(=O)NC(=O)NC1=O)C2=CCCCC2
|
|
| InChI |
InChI=1S/C12H16N2O3/c1-2-12(8-6-4-3-5-7-8)9(15)13-11(17)14-10(12)16/h6H,2-5,7H2,1H3,(H2,13,14,15,16,17)
|
|
| InChIKey |
WTYGAUXICFETTC-UHFFFAOYSA-N
|
|
| Synonyms |
CYCLOBARBITAL; Cyclobarbitone; Hexemal; Ethylhexabital; Phanodorm; 52-31-3; Tetrahydrophenobarbital; Amnosed; Cavonyl; Cyclodorm; Fanodormo; Namuron; Palinum; Philodorm; Pralumin; Sonaform; Adorm; Irifan; Pro-Sonil; Cyclobarbitol; Cyclobarbiton; Cyklodorm; Phanodorn; Ciclobarbital; Hypnoval; 5-(1-Cyclohexen-1-yl)-5-ethylbarbituric acid; Cyclobarbital [INN]; Cyclohexenyl-ethyl barbituric acid; 5-Ethyl-5-cyclohexenylbarbituric acid; 5-(1-Cyclohexenyl)-5-ethylbarbituric acid; 5-(1-Cyclohexen-1-yl)-5-ethyl-2,4,6(1H,3H,5H)-pyrimidinetrione; 0M8A98AD9H; Cyclobarbitonum; Cyclohexal; 2,4,6-(1H,3H,5H)-Pyrimidinetrione, 5-(1-cyclohexen-1-yl)-5-ethyl-; Barbituric acid, 5-(1-cyclohexen-1-yl)-5-ethyl-; Cyclobarbital (INN); Fanodorm; Hexemalum; Praelumin; Sonoform; 2,4,6(1H,3H,5H)-Pyrimidinetrione, 5-(1-cyclohexen-1-yl)-5-ethyl-; Cyclobarbitalum; Ciclobarbital [INN-Spanish]; Cyclobarbitalum [INN-Latin]; Cyclobarbitone [BAN]; EINECS 200-138-3; UNII-0M8A98AD9H; Cyclobarbital [INN:BAN:NF]; Zyklohexenylaethylbarbitursaeure; Ciclobarbital (TN); 5-(cyclohexen-1-yl)-5-ethyl-1,3-diazinane-2,4,6-trione; CYCLOBARBITAL [MI]; BIDD:PXR0072; CYCLOBARBITAL [MART.]; SCHEMBL157224; CYCLOBARBITAL [WHO-DD]; CHEMBL268164; DTXSID9022865; SCHEMBL23533905; CHEBI:134957; ZINC5651565; DB13737; D07323; 5-CYCLOHEX-1-ENYL-5-ETHYL-BARBITURIC ACID; Q416966; 5-Cyclohexenyl-5-A currencythyl-barbitursA currencyure; 2-Ethyl-6-(furan-2-yl)-4,5-dihydropyridazin-3(2H)-one; 5-cyclohex-1-en-1-yl-5-ethylpyrimidine-2,4,6(1H,3H,5H)-trione
|
|
| CAS | 52-31-3 | |
| PubChem CID | 5838 | |
| ChEMBL ID | CHEMBL268164 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 236.27 | ALogp: | 1.8 |
| HBD: | 2 | HBA: | 3 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 75.3 | Aromatic Rings: | 2 |
| Heavy Atoms: | 17 | QED Weighted: | 0.566 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.999 | MDCK Permeability: | 0.00004150 |
| Pgp-inhibitor: | 0.013 | Pgp-substrate: | 0.036 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.006 |
| 30% Bioavailability (F30%): | 0.005 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.999 | Plasma Protein Binding (PPB): | 54.45% |
| Volume Distribution (VD): | 0.711 | Fu: | 43.09% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.043 | CYP1A2-substrate: | 0.655 |
| CYP2C19-inhibitor: | 0.199 | CYP2C19-substrate: | 0.934 |
| CYP2C9-inhibitor: | 0.101 | CYP2C9-substrate: | 0.908 |
| CYP2D6-inhibitor: | 0.008 | CYP2D6-substrate: | 0.11 |
| CYP3A4-inhibitor: | 0.011 | CYP3A4-substrate: | 0.118 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 0.024 | Half-life (T1/2): | 0.694 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.002 | Human Hepatotoxicity (H-HT): | 0.102 |
| Drug-inuced Liver Injury (DILI): | 0.179 | AMES Toxicity: | 0.898 |
| Rat Oral Acute Toxicity: | 0.848 | Maximum Recommended Daily Dose: | 0.127 |
| Skin Sensitization: | 0.029 | Carcinogencity: | 0.072 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.007 |
| Respiratory Toxicity: | 0.9 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
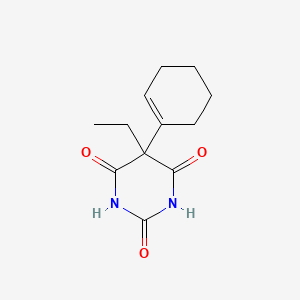
| ENC000698 | 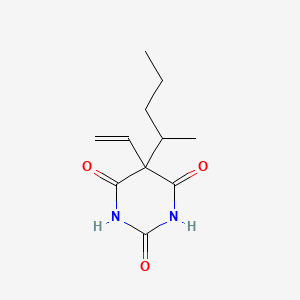 |
0.348 | D03WAJ | 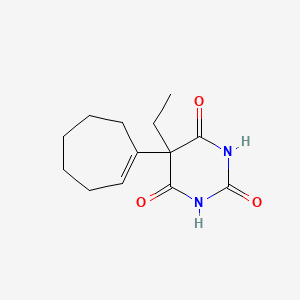 |
0.940 | ||
| ENC001339 | 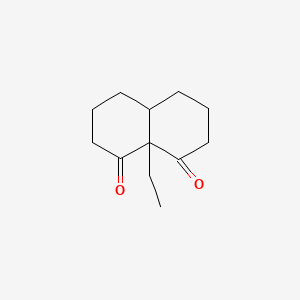 |
0.265 | D00ETS |  |
0.500 | ||
| ENC001318 | 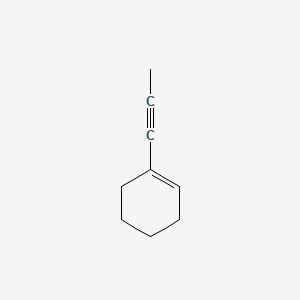 |
0.259 | D0Y7RW | 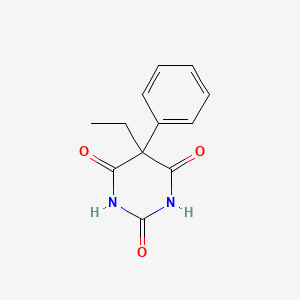 |
0.469 | ||
| ENC001229 | 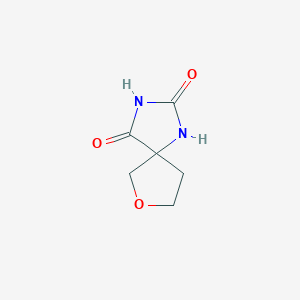 |
0.258 | D0E0WQ | 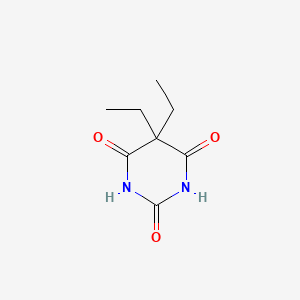 |
0.421 | ||
| ENC003728 | 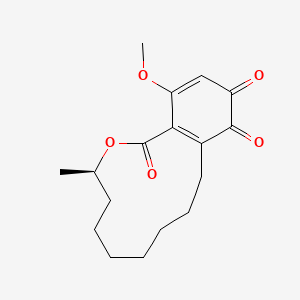 |
0.227 | D0A4JK | 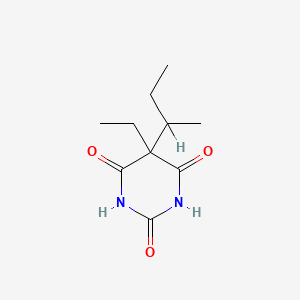 |
0.387 | ||
| ENC003479 | 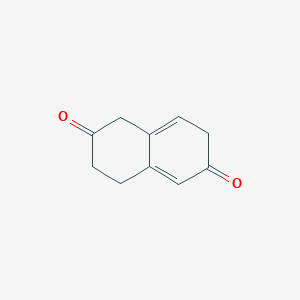 |
0.227 | D0O3AB | 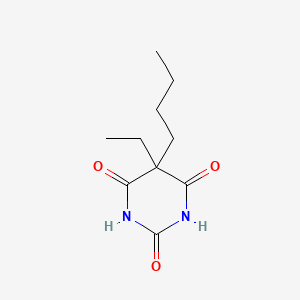 |
0.381 | ||
| ENC005740 | 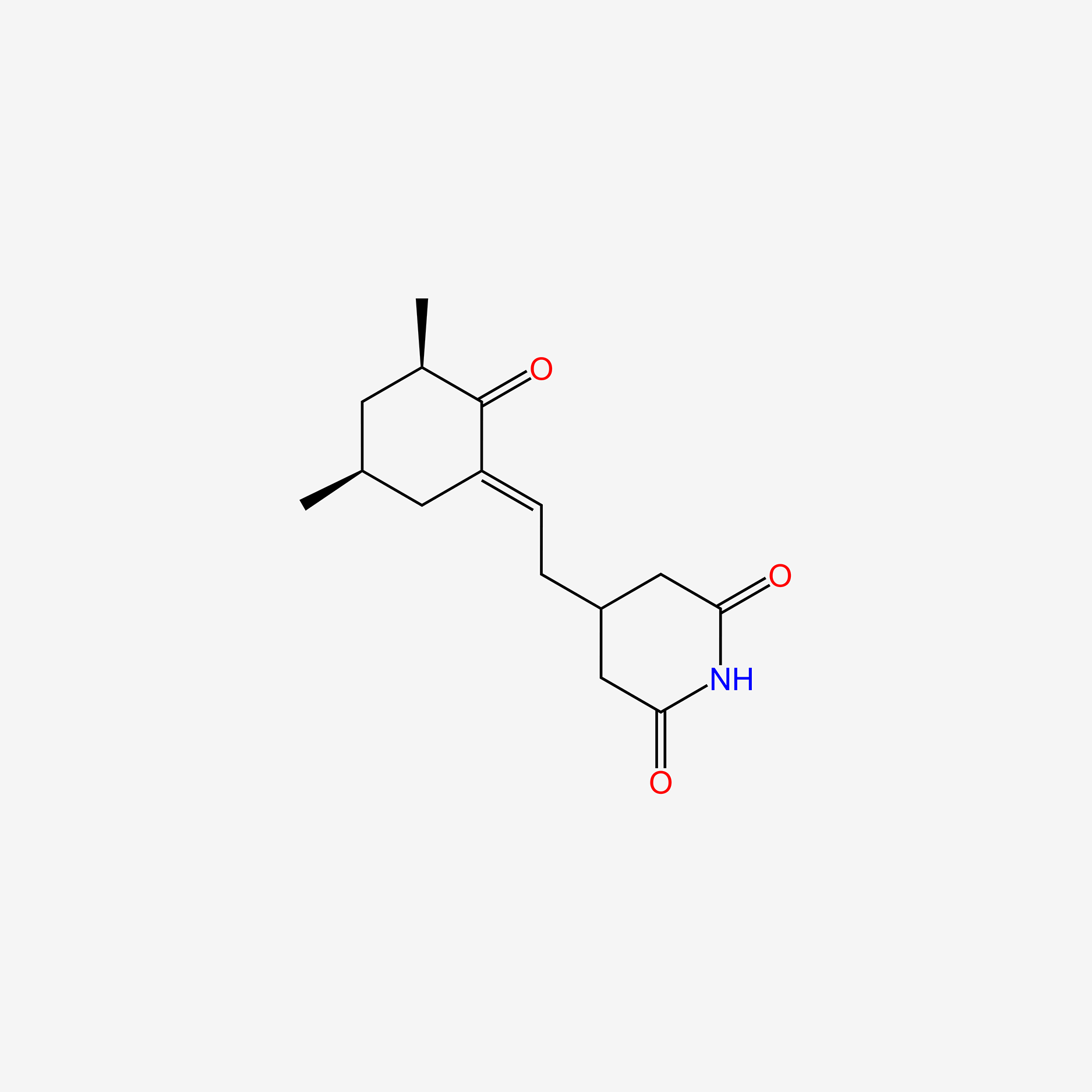 |
0.222 | D0R6BR | 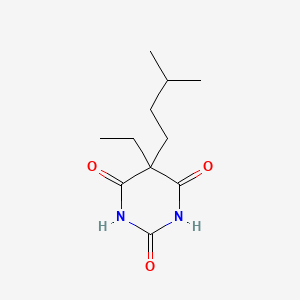 |
0.369 | ||
| ENC005741 | 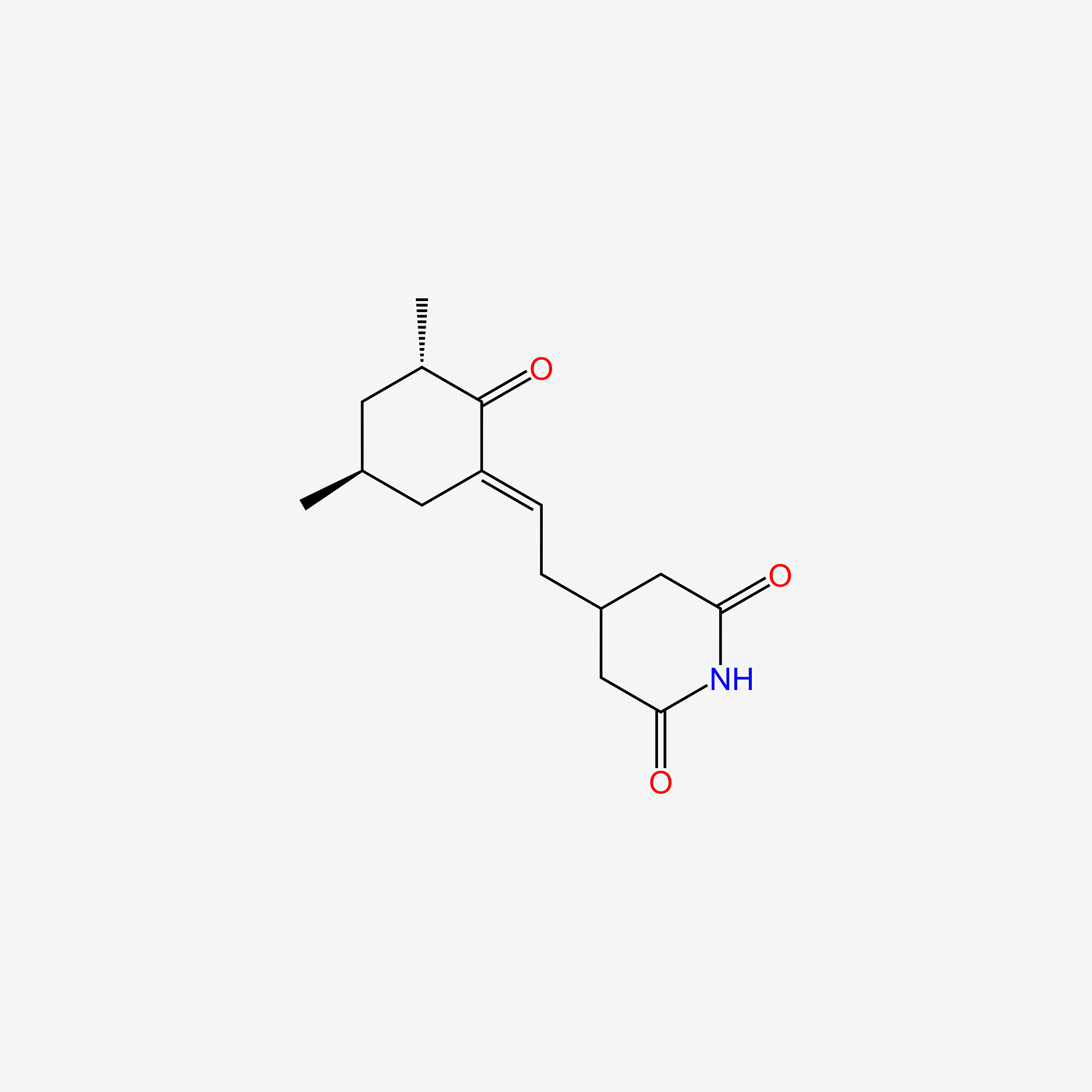 |
0.222 | D0F0YZ | 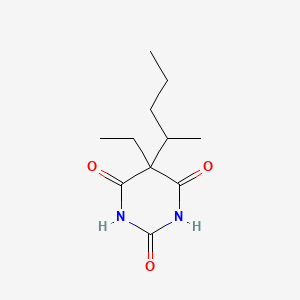 |
0.369 | ||
| ENC000450 | 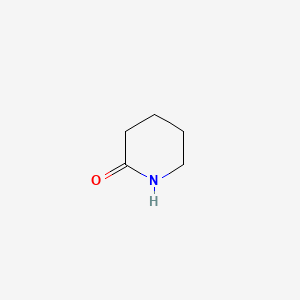 |
0.218 | D0W0MF | 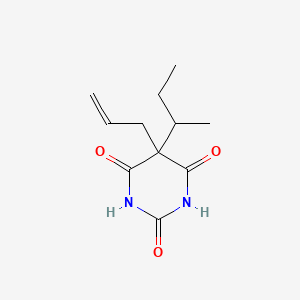 |
0.348 | ||
| ENC000965 | 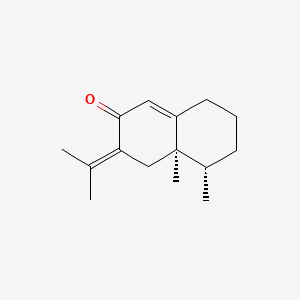 |
0.216 | D05TMQ | 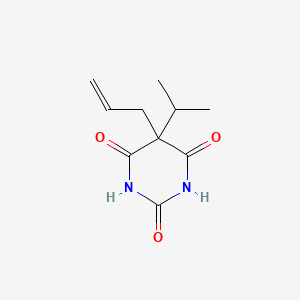 |
0.344 | ||