NPs Basic Information
|
Name |
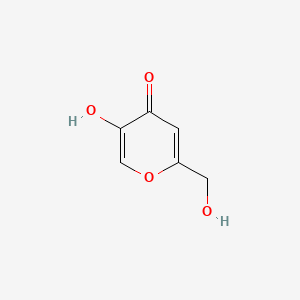
Kojic acid
|
| Molecular Formula | C6H6O4 | |
| IUPAC Name* |
5-hydroxy-2-(hydroxymethyl)pyran-4-one
|
|
| SMILES |
C1=C(OC=C(C1=O)O)CO
|
|
| InChI |
InChI=1S/C6H6O4/c7-2-4-1-5(8)6(9)3-10-4/h1,3,7,9H,2H2
|
|
| InChIKey |
BEJNERDRQOWKJM-UHFFFAOYSA-N
|
|
| Synonyms |
kojic acid; 501-30-4; 5-Hydroxy-2-(hydroxymethyl)-4H-pyran-4-one; 5-Hydroxy-2-(hydroxymethyl)-4-pyrone; 5-hydroxy-2-(hydroxymethyl)pyran-4-one; 5-Hydroxy-2-hydroxymethyl-4-pyrone; 4H-PYRAN-4-ONE, 5-HYDROXY-2-(HYDROXYMETHYL)-; acido kojico; 2-Hydroxymethyl-5-hydroxy-gamma-pyrone; 2-(Hydroxymethyl)-5-hydroxy-4H-pyran-4-one; MFCD00006580; 5-Hydroxy-2-hydroxymethyl-4H-4-pyranone; 5-Hydroxy-2-hydroxymethyl-pyran-4-one; 5-Hydroxy-2-hydroxymethyl-4H-pyran-4-one; Pyran-4-one, 5-hydroxy-2-(hydroxymethyl); CHEMBL287556; 123712-78-7; CHEBI:43572; NSC1942; 6K23F1TT52; NSC-1942; NCGC00017325-03; NCGC00017325-06; DSSTox_CID_20236; DSSTox_RID_79455; DSSTox_GSID_40236; CAS-501-30-4; CCRIS 4131; NSC 1942; EINECS 207-922-4; BRN 0120895; Kojisaeure; UNII-6K23F1TT52; AI3-02549; HSDB 7664; Spectrum_000191; KOJIC ACID [MI]; Spectrum2_001828; Spectrum3_001704; Spectrum4_000571; Spectrum5_001085; Natural Kojic Acid Powder; KOJIC ACID [HSDB]; KOJIC ACID [IARC]; KOJIC ACID [INCI]; Kojic Acid Dipalmitate 5kg; KOJIC ACID [MART.]; Oprea1_038773; SCHEMBL36895; BSPBio_003288; KBioGR_001002; KBioSS_000671; KOJIC ACID [WHO-DD]; 5-18-02-00516 (Beilstein Handbook Reference); BIDD:ER0501; DivK1c_000923; SPBio_001875; Kojic acid, analytical standard; MEGxm0_000388; WLN: T6O DVJ B1Q EQ; DTXSID2040236; ACon1_000622; BEJNERDRQOWKJM-UHFFFAOYSA-; HMS502O05; KBio1_000923; KBio2_000671; KBio2_003239; KBio2_005807; KBio3_002508; NINDS_000923; HMS3604L04; KUC106760N; TNP00261; 2-Hydroxymethyl-5-hydroxy-?-pyrone; 2-Hydroxymethyl-5-Hydroxy-G-Pyrone; Tox21_110814; Tox21_113449; BBL010645; BDBM50031467; CCG-38458; s5174; STK801688; ZINC13831818; AKOS000120649; Tox21_110814_1; 5-Hydroxy-2-hydroxymethyl-I(3)-pyron; DB01759; HY-W050154; IDI1_000923; SMP1_000171; NCGC00017325-01; NCGC00017325-02; NCGC00017325-04; NCGC00017325-05; NCGC00017325-10; NCGC00142361-01; NCGC00168903-01; NCGC00168903-02; NCGC00181145-01; AC-11658; AS-11648; Kojic acid, purum, >=98.0% (HPLC); KSC-11-228-2; SY018009; 2-HYDROXYMETHYL-5-HYDROXY-4-PYRONE; 2-Hydroxymethyl-5-hydroxy-4H-pyran-4-one; CS-0032701; FT-0627581; K0010; K0012; EN300-20919; 01K304; C91105; K-7000; 2-HYDROXYMETHYL-5-HYDROXY-.GAMMA.-PYRONE; Q416285; SR-01000945134; SR-01000945134-1; C65A72E0-0E46-4AF4-AA68-178ECA2E5FCC; F0001-1307; Z104484860
|
|
| CAS | 501-30-4 | |
| PubChem CID | 3840 | |
| ChEMBL ID | CHEMBL287556 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 142.11 | ALogp: | -0.9 |
| HBD: | 2 | HBA: | 4 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 66.8 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.589 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.591 | MDCK Permeability: | 0.00006400 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.348 |
| Human Intestinal Absorption (HIA): | 0.013 | 20% Bioavailability (F20%): | 0.176 |
| 30% Bioavailability (F30%): | 0.93 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.082 | Plasma Protein Binding (PPB): | 53.66% |
| Volume Distribution (VD): | 0.665 | Fu: | 69.76% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.236 | CYP1A2-substrate: | 0.137 |
| CYP2C19-inhibitor: | 0.036 | CYP2C19-substrate: | 0.064 |
| CYP2C9-inhibitor: | 0.012 | CYP2C9-substrate: | 0.476 |
| CYP2D6-inhibitor: | 0.01 | CYP2D6-substrate: | 0.505 |
| CYP3A4-inhibitor: | 0.007 | CYP3A4-substrate: | 0.128 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.066 | Half-life (T1/2): | 0.888 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.058 | Human Hepatotoxicity (H-HT): | 0.11 |
| Drug-inuced Liver Injury (DILI): | 0.387 | AMES Toxicity: | 0.028 |
| Rat Oral Acute Toxicity: | 0.2 | Maximum Recommended Daily Dose: | 0.013 |
| Skin Sensitization: | 0.535 | Carcinogencity: | 0.722 |
| Eye Corrosion: | 0.019 | Eye Irritation: | 0.884 |
| Respiratory Toxicity: | 0.165 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC006095 | 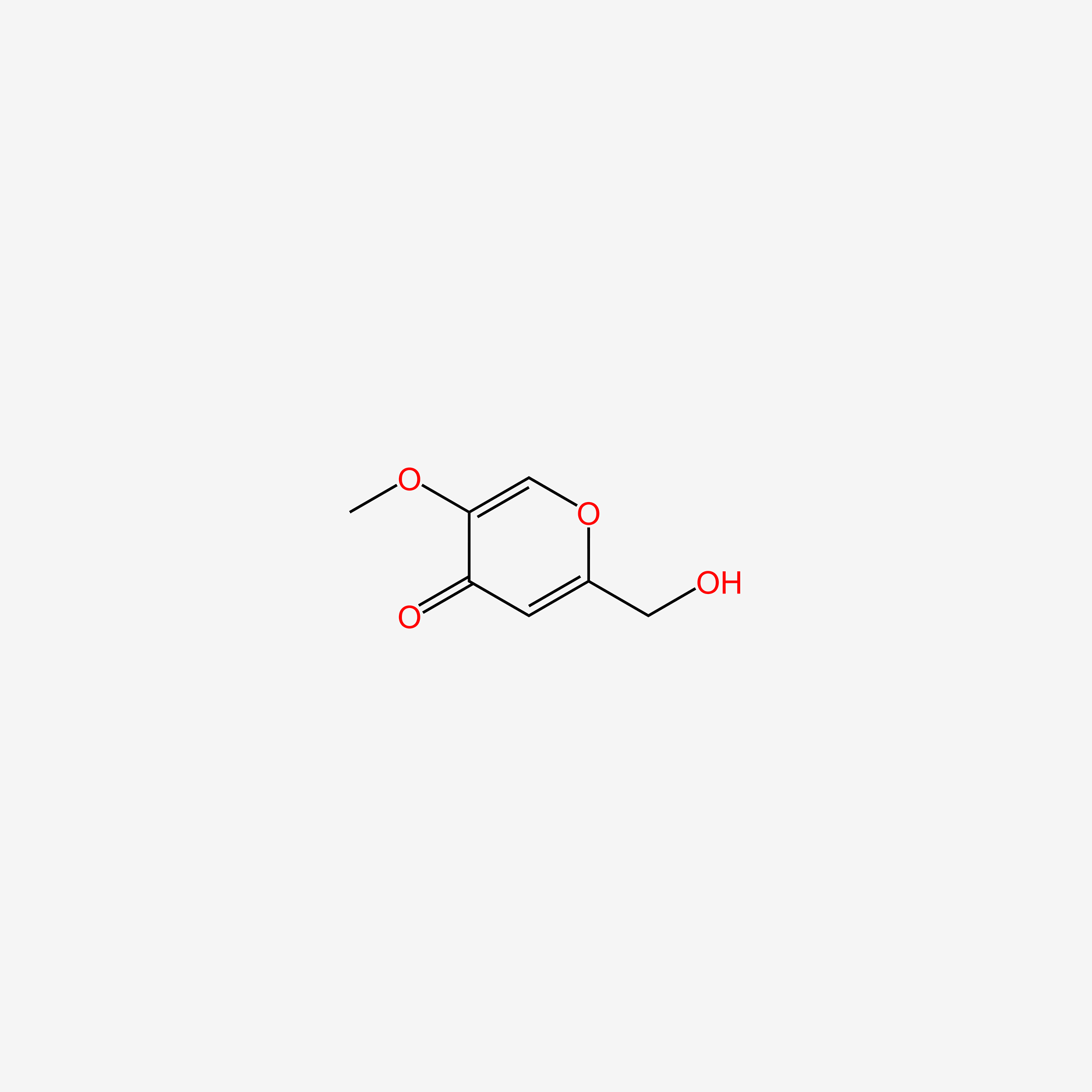 |
0.583 | D01WJL | 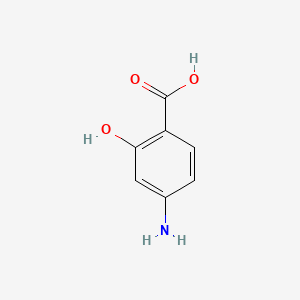 |
0.244 | ||
| ENC002506 | 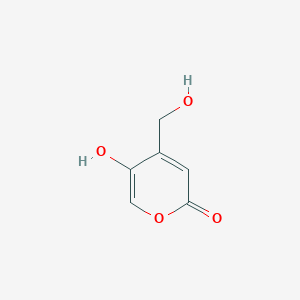 |
0.543 | D0C4YC | 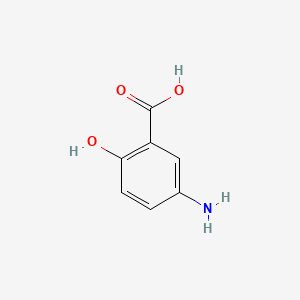 |
0.244 | ||
| ENC002334 | 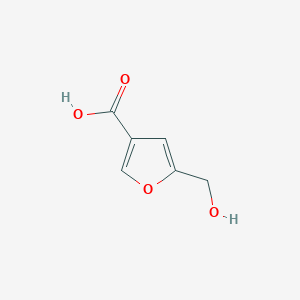 |
0.500 | D0T7OW | 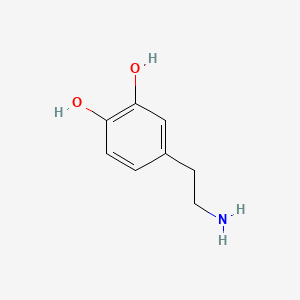 |
0.239 | ||
| ENC005612 | 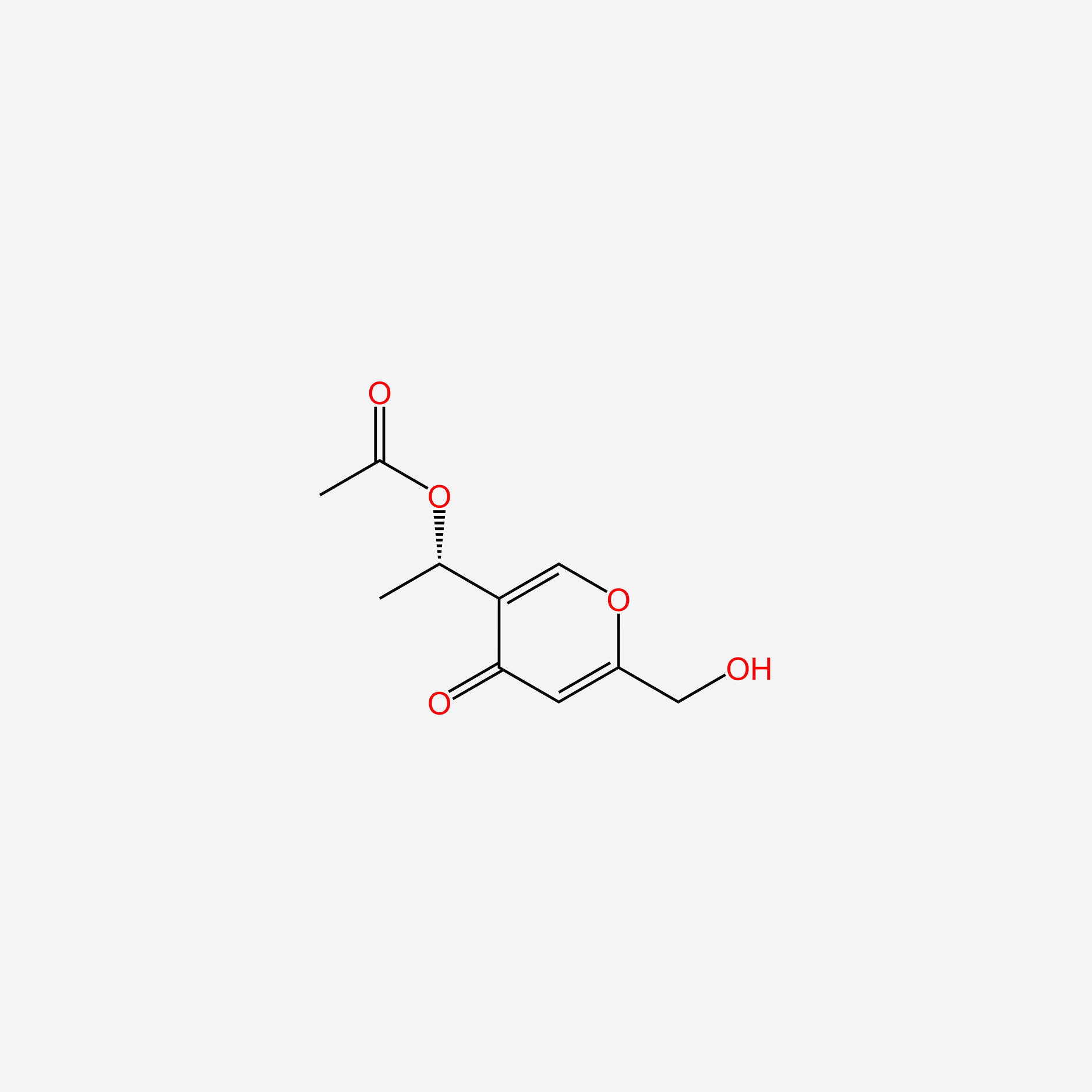 |
0.457 | D07AHW | 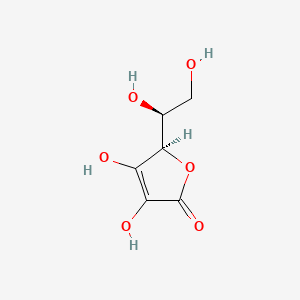 |
0.234 | ||
| ENC006096 | 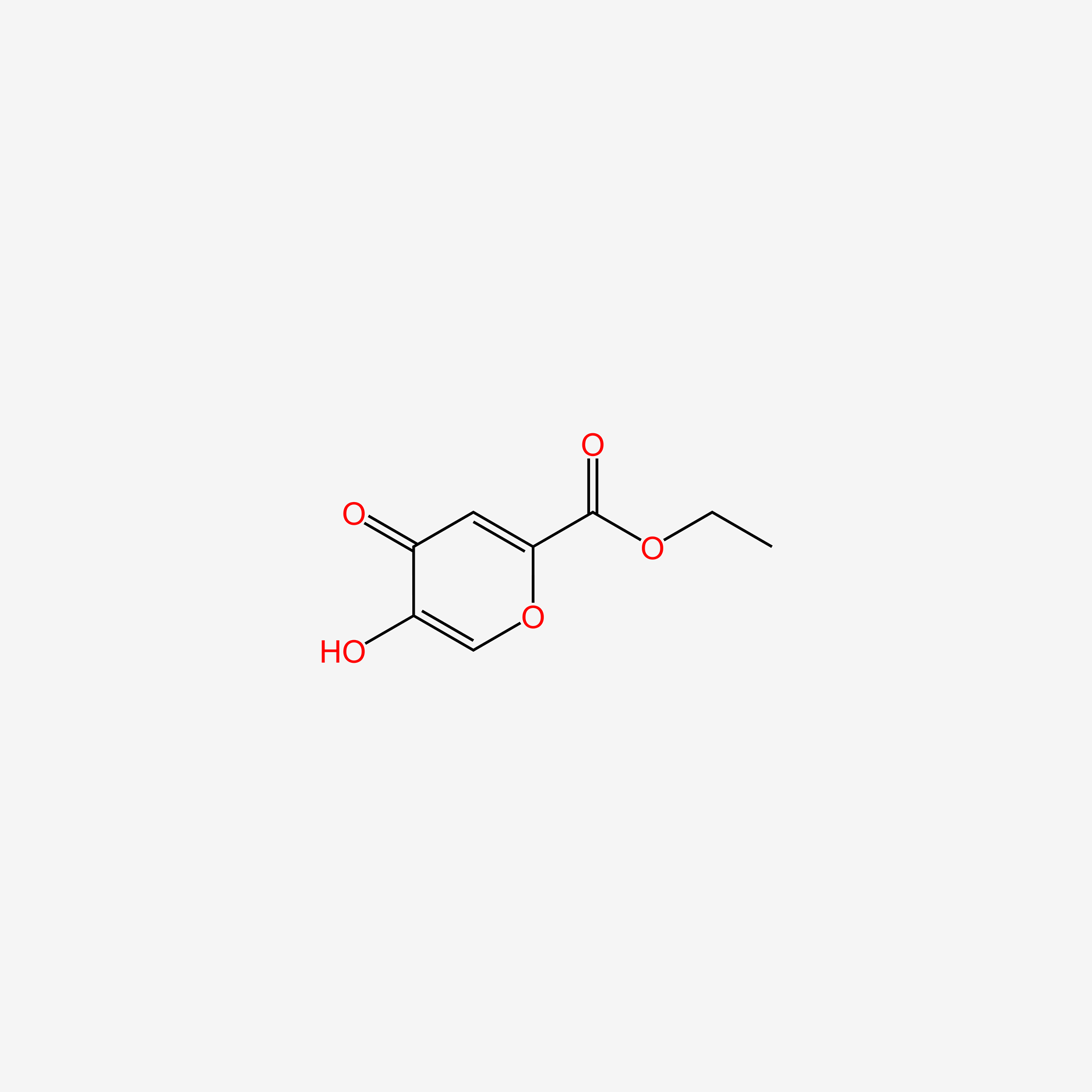 |
0.442 | D0N0OU | 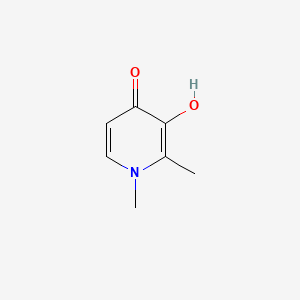 |
0.233 | ||
| ENC004401 |  |
0.392 | D08HVR | 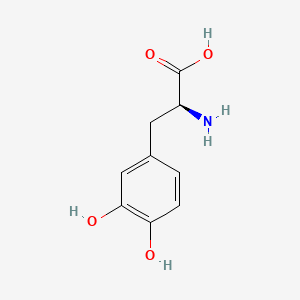 |
0.231 | ||
| ENC002730 | 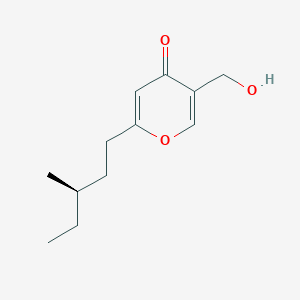 |
0.388 | D07MOX | 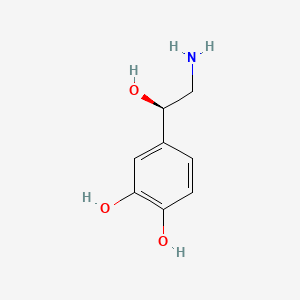 |
0.229 | ||
| ENC001951 | 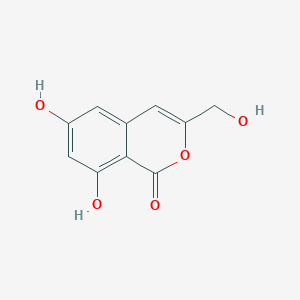 |
0.360 | D0BA6T | 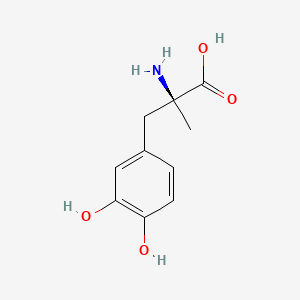 |
0.222 | ||
| ENC004766 | 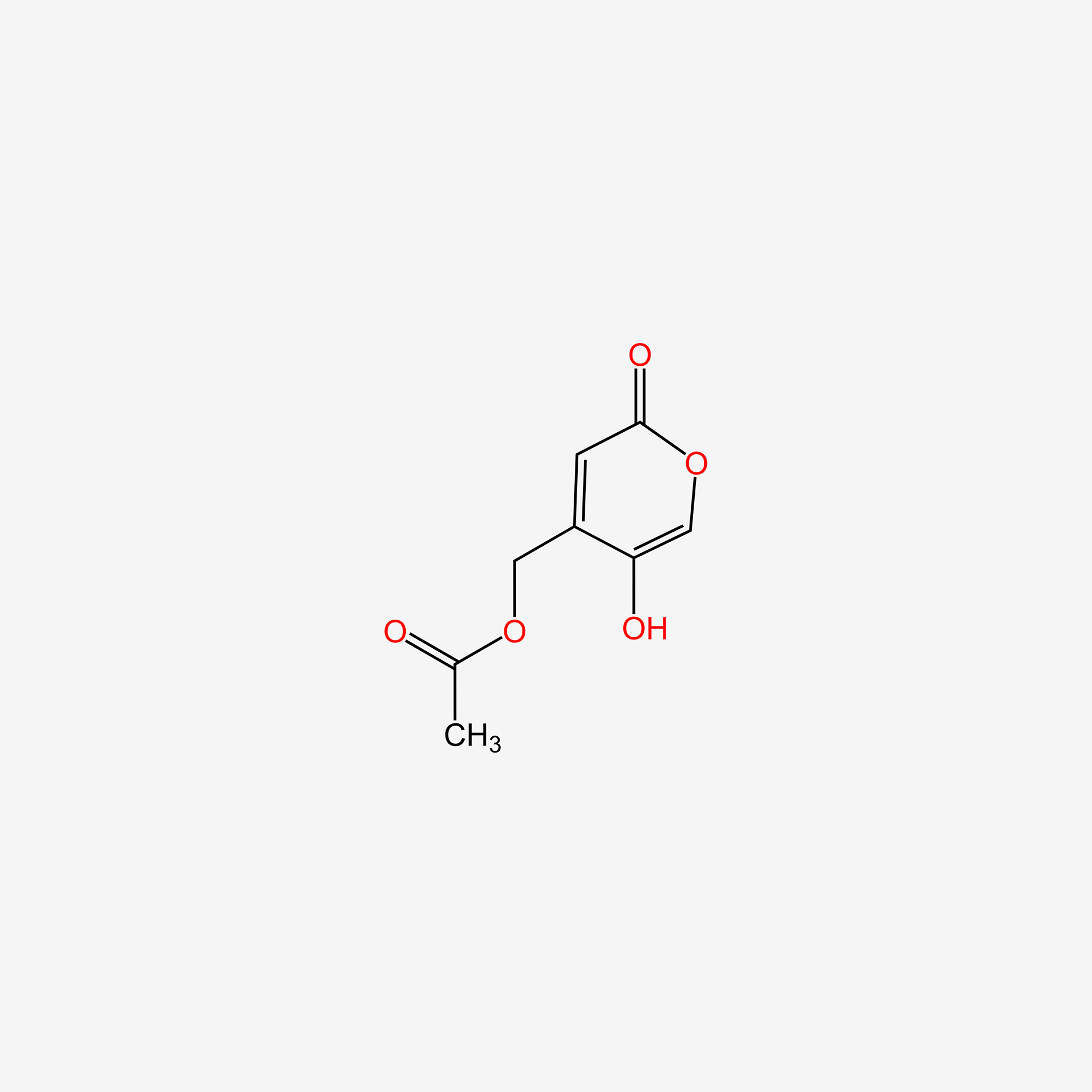 |
0.348 | D02ZJI | 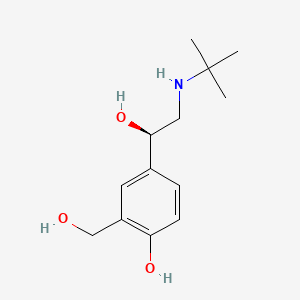 |
0.220 | ||
| ENC005611 | 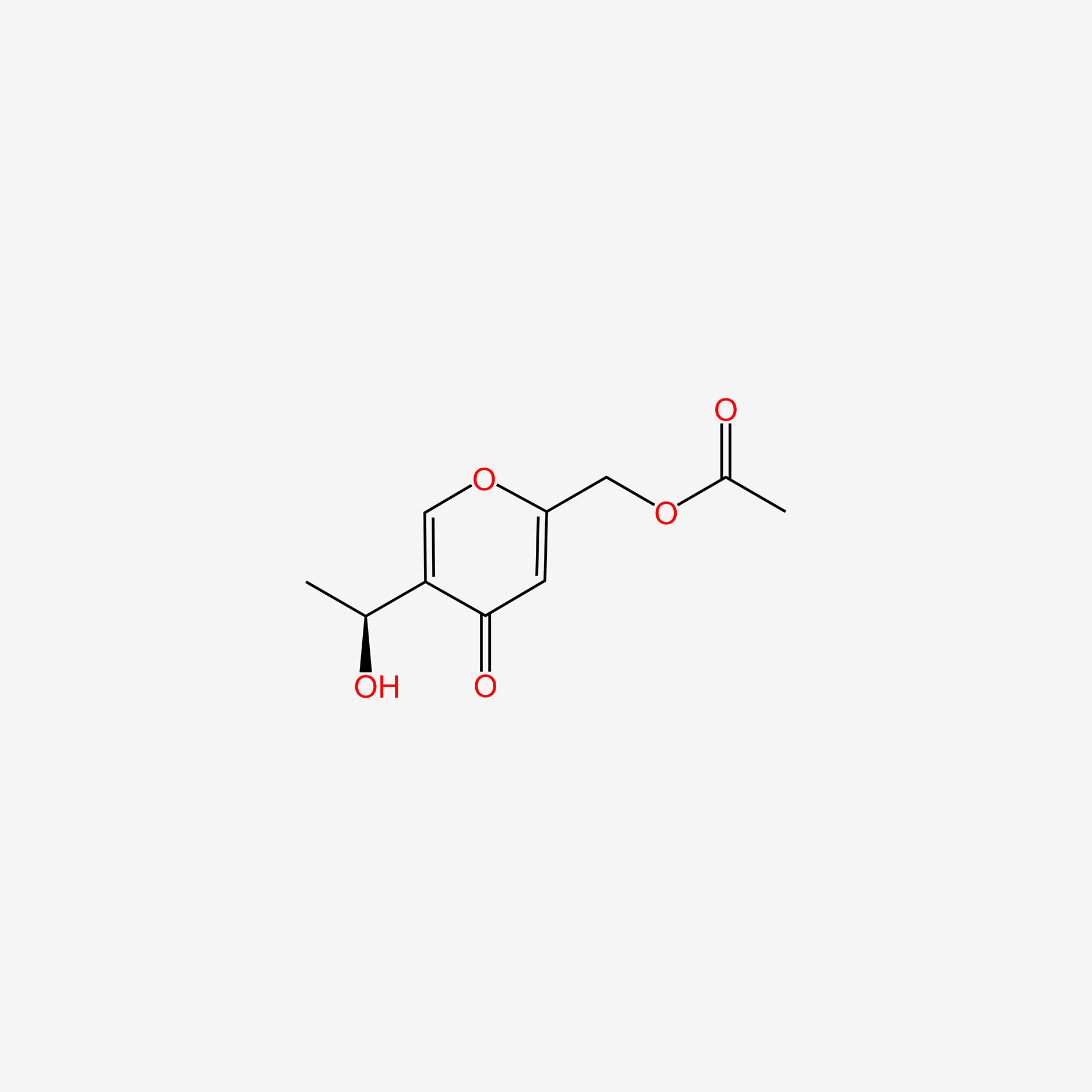 |
0.340 | D0K5CB | 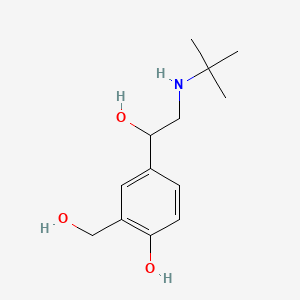 |
0.220 | ||