NPs Basic Information
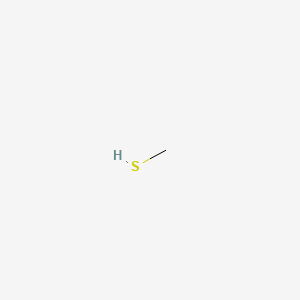
|
Name |
Methanethiol
|
| Molecular Formula | CH4S | |
| IUPAC Name* |
methanethiol
|
|
| SMILES |
CS
|
|
| InChI |
InChI=1S/CH4S/c1-2/h2H,1H3
|
|
| InChIKey |
LSDPWZHWYPCBBB-UHFFFAOYSA-N
|
|
| Synonyms |
methanethiol; METHYL MERCAPTAN; Methylmercaptan; Mercaptomethane; 74-93-1; Methyl sulfhydrate; Thiomethanol; Methanthiol; Thiomethyl alcohol; Metilmercaptano; Methvtiolo; Methylmercaptaan; Mercaptan methylique; Methaanthiol; Thiomethane; RCRA waste number U153; FEMA No. 2716; Methanethiole; CH3SH; methyl-mercaptan; Methyl thioalcohol; MeSH; UN 1064; 2X8406WW9I; Methaanthiol [Dutch]; Methanthiol [German]; Methvtiolo [Italian]; Methylmercaptaan [Dutch]; Metilmercaptano [Italian]; Metilmercaptano [Spanish]; Methyl mercaptan (natural); Mercaptan methylique [French]; HSDB 813; EINECS 200-822-1; UN1064; RCRA waste no. U153; BRN 1696840; methane thiol; methyl sulfides; methyl thiol; methyl-thiol; UNII-2X8406WW9I; (methyl)sulfane; Methylthioalcohol; a methyl thioether; sulfonium methylide; Methanethiol, purum; Methanethiol, 98.0%; METHANETHIOL [MI]; EC 200-822-1; Methanethiol, >=98.0%; 4-01-00-01273 (Beilstein Handbook Reference); METHYL MERCAPTAN [FHFI]; METHYL MERCAPTAN [HSDB]; DTXSID5026382; CHEBI:16007; CHEBI:86315; NSC229573; AKOS009157032; NSC-229573; Methyl mercaptan [UN1064] [Poison gas]; C00409; Q409309; 17719-48-1; Z22
|
|
| CAS | 74-93-1 | |
| PubChem CID | 878 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 48.11 | ALogp: | 0.5 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 1.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 2 | QED Weighted: | 0.387 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.5 | MDCK Permeability: | 0.00001510 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.01 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.048 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.846 | Plasma Protein Binding (PPB): | 55.57% |
| Volume Distribution (VD): | 1.393 | Fu: | 64.81% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.515 | CYP1A2-substrate: | 0.717 |
| CYP2C19-inhibitor: | 0.043 | CYP2C19-substrate: | 0.85 |
| CYP2C9-inhibitor: | 0.005 | CYP2C9-substrate: | 0.218 |
| CYP2D6-inhibitor: | 0.005 | CYP2D6-substrate: | 0.542 |
| CYP3A4-inhibitor: | 0.005 | CYP3A4-substrate: | 0.26 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.517 | Half-life (T1/2): | 0.804 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.005 | Human Hepatotoxicity (H-HT): | 0.358 |
| Drug-inuced Liver Injury (DILI): | 0.288 | AMES Toxicity: | 0.136 |
| Rat Oral Acute Toxicity: | 0.83 | Maximum Recommended Daily Dose: | 0.037 |
| Skin Sensitization: | 0.394 | Carcinogencity: | 0.794 |
| Eye Corrosion: | 0.977 | Eye Irritation: | 0.994 |
| Respiratory Toxicity: | 0.94 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000046 |  |
0.200 | D05KEZ |  |
0.125 | ||
| ENC000039 |  |
0.125 | D00AMQ | 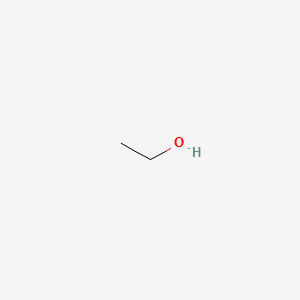 |
0.125 | ||
| ENC000286 | 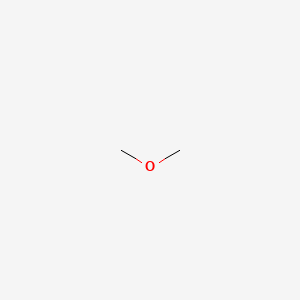 |
0.125 | D04CRL | 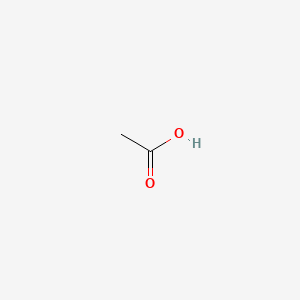 |
0.100 | ||
| ENC000689 | 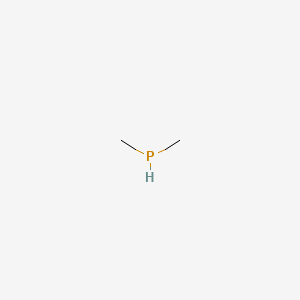 |
0.125 | D08HVE | 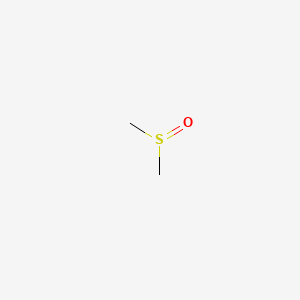 |
0.100 | ||
| ENC000008 | 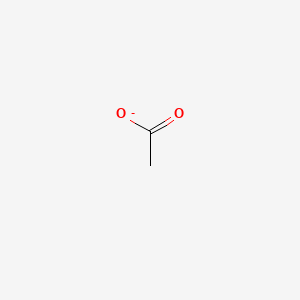 |
0.100 | D08HZC | 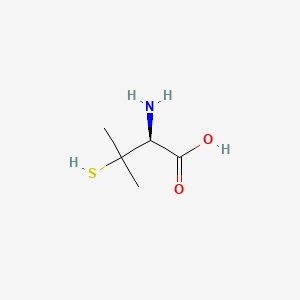 |
0.091 | ||
| ENC000009 | 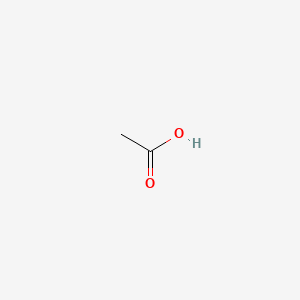 |
0.100 | D0V0LB | 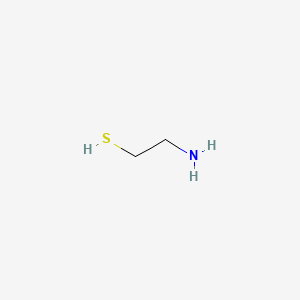 |
0.091 | ||
| ENC000417 | 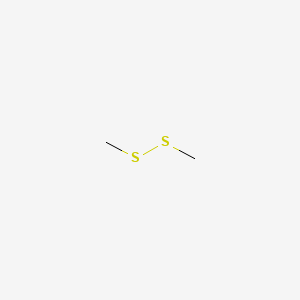 |
0.091 | D0Z4UY | 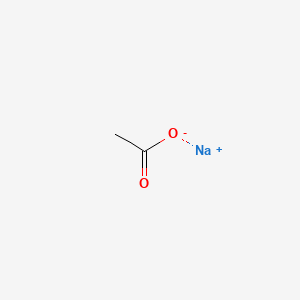 |
0.091 | ||
| ENC000469 | 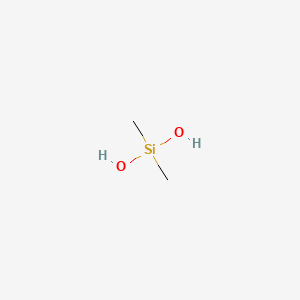 |
0.083 | D0C1PY | 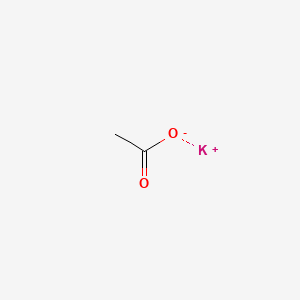 |
0.091 | ||
| ENC000465 | 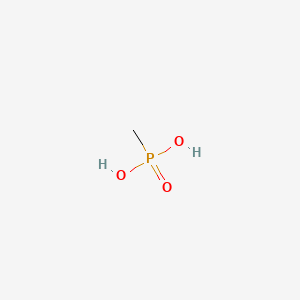 |
0.083 | D06XGW | 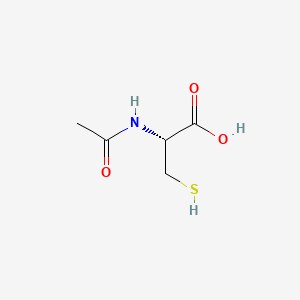 |
0.077 | ||
| ENC000132 | 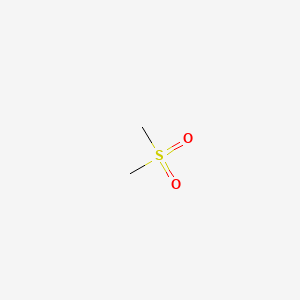 |
0.083 | D0A8CJ | 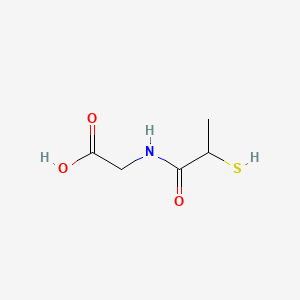 |
0.077 | ||