NPs Basic Information
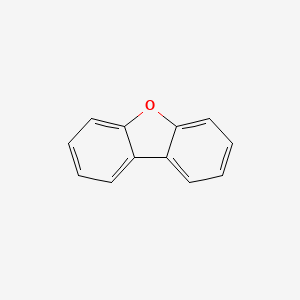
|
Name |
Dibenzofuran
|
| Molecular Formula | C12H8O | |
| IUPAC Name* |
dibenzofuran
|
|
| SMILES |
C1=CC=C2C(=C1)C3=CC=CC=C3O2
|
|
| InChI |
InChI=1S/C12H8O/c1-3-7-11-9(5-1)10-6-2-4-8-12(10)13-11/h1-8H
|
|
| InChIKey |
TXCDCPKCNAJMEE-UHFFFAOYSA-N
|
|
| Synonyms |
dibenzofuran; Dibenzo[b,d]furan; 132-64-9; diphenylene oxide; Dibenzofurans; 2,2'-Biphenylene oxide; 2,2'-Biphenylylene oxide; Dibenzo(b,d)furan; dibenzofurane; (1,1'-Biphenyl)-2,2'-diyl oxide; [1,1'-Biphenyl]-2,2'-diyl oxide; CHEBI:28145; 8U54U639VI; NSC-1245; DSSTox_CID_1993; DSSTox_RID_76446; DSSTox_GSID_21993; Dibenzol(b,d)furan; 102250-99-7; CAS-132-64-9; CCRIS 1436; HSDB 2163; NSC 1245; EINECS 205-071-3; UNII-8U54U639VI; AI3-00039; Dibenzo[b]furan; dibenzo[bd]furan; Dibenzofuran, 98%; [1,2'-diyl oxide; DIBENZOFURAN [MI]; bmse000548; DIBENZOFURAN [HSDB]; SCHEMBL8207; CHEMBL277497; DTXSID2021993; Dibenzofuran, analytical standard; NSC1245; DIBENZO (b,d) FURAN (purity); ZINC3861058; Tox21_202116; Tox21_300052; BDBM50408362; MFCD00004968; STL185574; AKOS000120971; CS-W017802; PS-5378; NCGC00164102-01; NCGC00164102-02; NCGC00164102-03; NCGC00254221-01; NCGC00259665-01; AC-19766; DB-042123; D0147; FT-0624634; EN300-18022; C07729; Q419513; Q-101160; Z57127512; Dibenzo[b,d]furan, BCR(R) certified Reference Material; 8-oxatricyclo[7.4.0.0,2,7]trideca-1(9),2(7),3,5,10,12-hexaene; 8-oxatricyclo[7.4.0.0^{2,7}]trideca-1(9),2(7),3,5,10,12-hexaene; 1IT
|
|
| CAS | 132-64-9 | |
| PubChem CID | 568 | |
| ChEMBL ID | CHEMBL277497 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 168.19 | ALogp: | 4.1 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 13.1 | Aromatic Rings: | 3 |
| Heavy Atoms: | 13 | QED Weighted: | 0.487 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.611 | MDCK Permeability: | 0.00001950 |
| Pgp-inhibitor: | 0.035 | Pgp-substrate: | 0.16 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.996 |
| 30% Bioavailability (F30%): | 0.994 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.266 | Plasma Protein Binding (PPB): | 95.99% |
| Volume Distribution (VD): | 1.03 | Fu: | 4.38% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.995 | CYP1A2-substrate: | 0.648 |
| CYP2C19-inhibitor: | 0.825 | CYP2C19-substrate: | 0.388 |
| CYP2C9-inhibitor: | 0.361 | CYP2C9-substrate: | 0.868 |
| CYP2D6-inhibitor: | 0.433 | CYP2D6-substrate: | 0.906 |
| CYP3A4-inhibitor: | 0.113 | CYP3A4-substrate: | 0.242 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 8.014 | Half-life (T1/2): | 0.221 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.093 | Human Hepatotoxicity (H-HT): | 0.154 |
| Drug-inuced Liver Injury (DILI): | 0.915 | AMES Toxicity: | 0.81 |
| Rat Oral Acute Toxicity: | 0.251 | Maximum Recommended Daily Dose: | 0.112 |
| Skin Sensitization: | 0.776 | Carcinogencity: | 0.87 |
| Eye Corrosion: | 0.434 | Eye Irritation: | 0.995 |
| Respiratory Toxicity: | 0.693 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000047 | 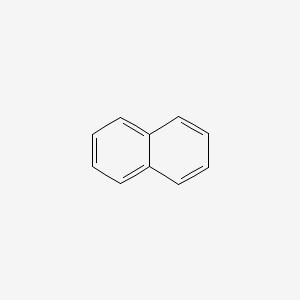 |
0.468 | D02TJS | 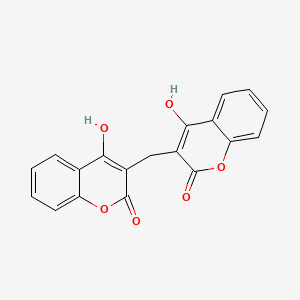 |
0.486 | ||
| ENC000171 | 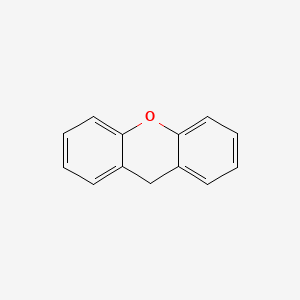 |
0.446 | D0QV5T | 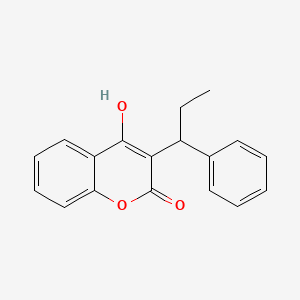 |
0.456 | ||
| ENC000159 | 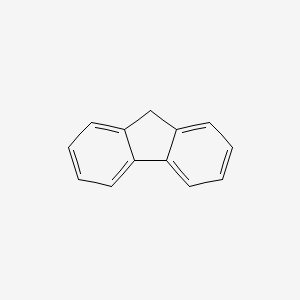 |
0.444 | D0B1FE | 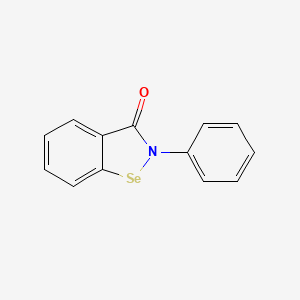 |
0.433 | ||
| ENC002806 | 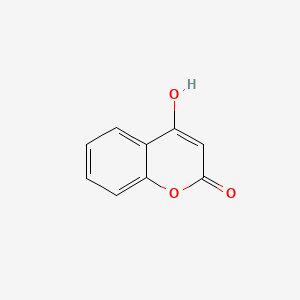 |
0.431 | D0E3OF | 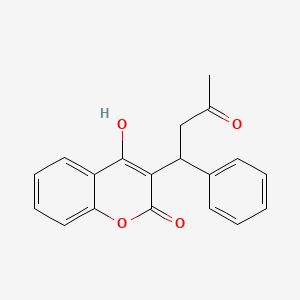 |
0.425 | ||
| ENC000737 | 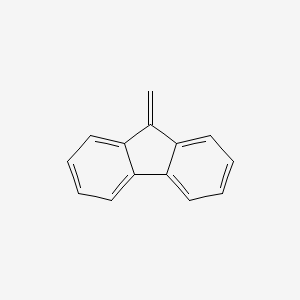 |
0.429 | D08FTG | 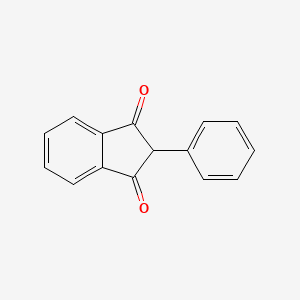 |
0.375 | ||
| ENC000167 | 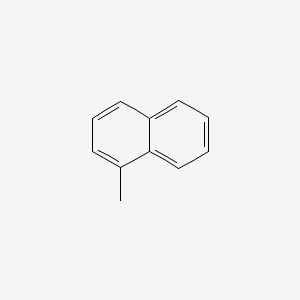 |
0.420 | D04MSM |  |
0.358 | ||
| ENC000025 |  |
0.420 | D0Y0JH |  |
0.353 | ||
| ENC000321 |  |
0.408 | D04QZD | 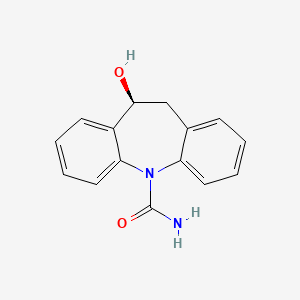 |
0.348 | ||
| ENC002323 | 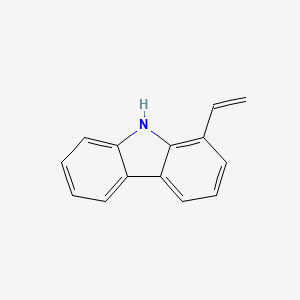 |
0.407 | D0QL3P | 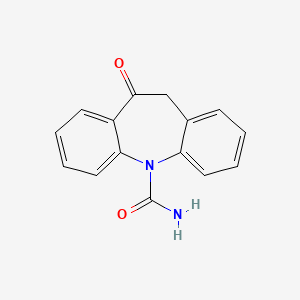 |
0.348 | ||
| ENC000732 | 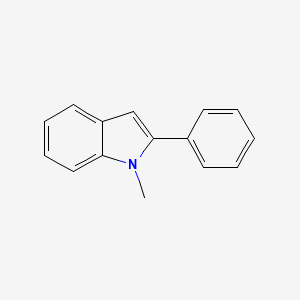 |
0.387 | D0G1VX |  |
0.344 | ||