NPs Basic Information
|
Name |
Fluorene
|
| Molecular Formula | C13H10 | |
| IUPAC Name* |
9H-fluorene
|
|
| SMILES |
C1C2=CC=CC=C2C3=CC=CC=C31
|
|
| InChI |
InChI=1S/C13H10/c1-3-7-12-10(5-1)9-11-6-2-4-8-13(11)12/h1-8H,9H2
|
|
| InChIKey |
NIHNNTQXNPWCJQ-UHFFFAOYSA-N
|
|
| Synonyms |
FLUORENE; 9H-Fluorene; 86-73-7; Diphenylenemethane; o-Biphenylenemethane; 2,3-Benzindene; 2,2'-Methylenebiphenyl; o-Biphenylmethane; Methane, diphenylene-; Fluoren; flourene; alpha-diphenylenemethane-9H-fluorene; CHEBI:28266; 3Q2UY0968A; NSC-6787; Fluorene 10 microg/mL in Cyclohexane; Fluorene 10 microg/mL in Acetonitrile; Fluorene 100 microg/mL in Acetonitrile; DSSTox_CID_4105; DSSTox_RID_77291; DSSTox_GSID_24105; 95270-88-5; CAS-86-73-7; CCRIS 947; HSDB 2165; NSC 6787; EINECS 201-695-5; MFCD00001111; UNII-3Q2UY0968A; AI3-09074; Fluorene, Reagent; Fluorene, 98%; FLUORENE [HSDB]; FLUORENE [IARC]; bmse000524; 9H-FLUORENE [MI]; EC 201-695-5; Fluorene, analytical standard; CHEMBL16236; ghl.PD_Mitscher_leg0.1322; DTXSID8024105; NSC6787; ZINC968333; Fluorene, >=99.0% (HPLC); ACT08954; AMY38998; STR04556; Tox21_202140; Tox21_300572; BBL027323; STK802351; AKOS000119854; AC-5810; HY-W026772; NCGC00164052-01; NCGC00164052-02; NCGC00164052-03; NCGC00254303-01; NCGC00259689-01; CS-0070796; F0017; F0061; FT-0626447; FT-0668575; EN300-18388; Fluorene Zone Refined (number of passes:70); C07715; Fluorene, Zone Refined (number of passes:70); Q417934; Q-201117; Fluorene, certified reference material, TraceCERT(R); Z57127470; F1313-0006; 9FL
|
|
| CAS | 86-73-7 | |
| PubChem CID | 6853 | |
| ChEMBL ID | CHEMBL16236 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 166.22 | ALogp: | 4.2 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 3 |
| Heavy Atoms: | 13 | QED Weighted: | 0.473 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.458 | MDCK Permeability: | 0.00002920 |
| Pgp-inhibitor: | 0.022 | Pgp-substrate: | 0.026 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.974 |
| 30% Bioavailability (F30%): | 0.019 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.713 | Plasma Protein Binding (PPB): | 96.88% |
| Volume Distribution (VD): | 1.177 | Fu: | 2.42% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.887 | CYP1A2-substrate: | 0.56 |
| CYP2C19-inhibitor: | 0.782 | CYP2C19-substrate: | 0.09 |
| CYP2C9-inhibitor: | 0.492 | CYP2C9-substrate: | 0.684 |
| CYP2D6-inhibitor: | 0.099 | CYP2D6-substrate: | 0.878 |
| CYP3A4-inhibitor: | 0.166 | CYP3A4-substrate: | 0.44 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.716 | Half-life (T1/2): | 0.176 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.045 | Human Hepatotoxicity (H-HT): | 0.189 |
| Drug-inuced Liver Injury (DILI): | 0.836 | AMES Toxicity: | 0.721 |
| Rat Oral Acute Toxicity: | 0.052 | Maximum Recommended Daily Dose: | 0.124 |
| Skin Sensitization: | 0.169 | Carcinogencity: | 0.768 |
| Eye Corrosion: | 0.036 | Eye Irritation: | 0.988 |
| Respiratory Toxicity: | 0.037 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
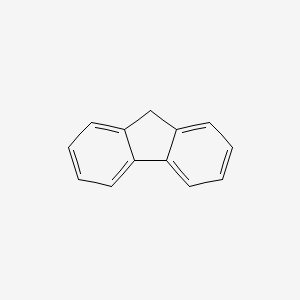
| ENC000171 | 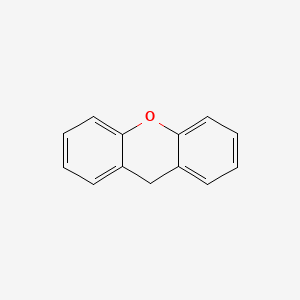 |
0.620 | D04WFD | 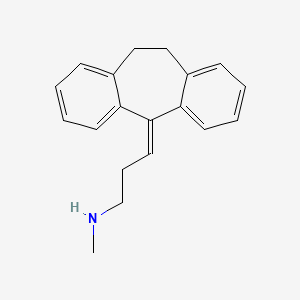 |
0.508 | ||
| ENC000737 |  |
0.538 | D0Y5UG | 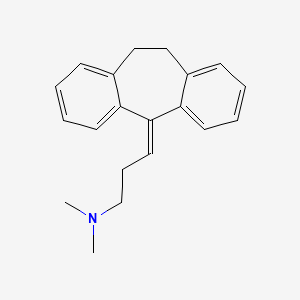 |
0.493 | ||
| ENC000047 | 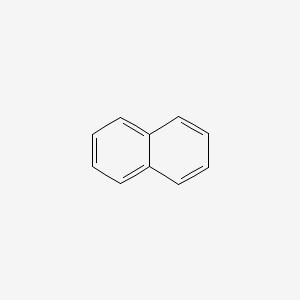 |
0.468 | D0DV3O | 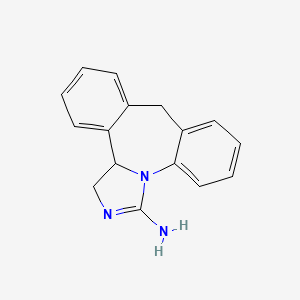 |
0.484 | ||
| ENC001372 | 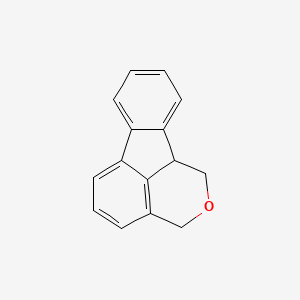 |
0.450 | D0QL3P | 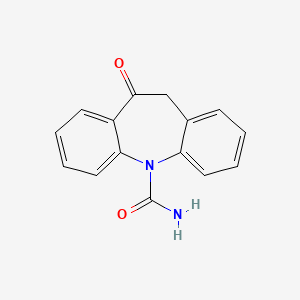 |
0.476 | ||
| ENC000036 | 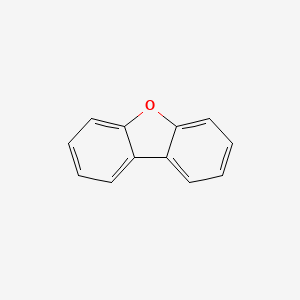 |
0.444 | D08FTG | 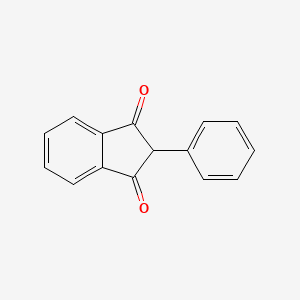 |
0.467 | ||
| ENC001371 | 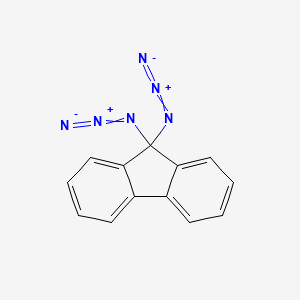 |
0.424 | D0R6RO | 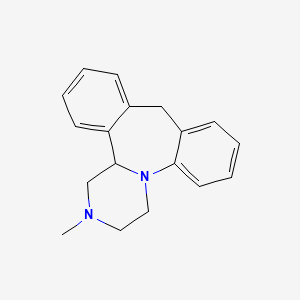 |
0.463 | ||
| ENC000321 | 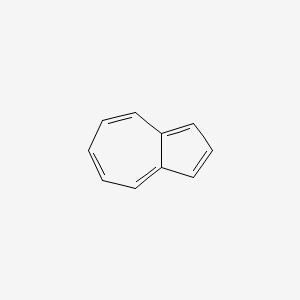 |
0.408 | D06FES | 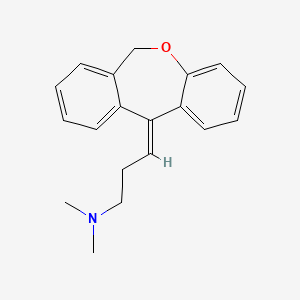 |
0.449 | ||
| ENC001375 | 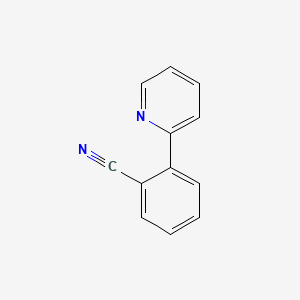 |
0.404 | D0Q2MN | 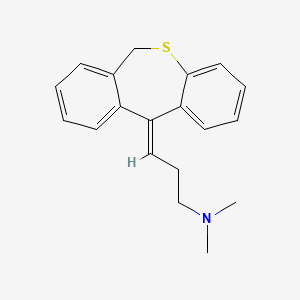 |
0.449 | ||
| ENC000732 | 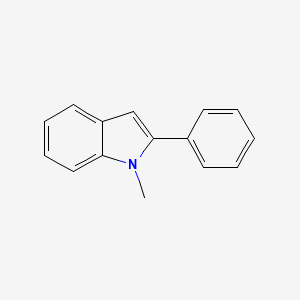 |
0.387 | D01UTL | 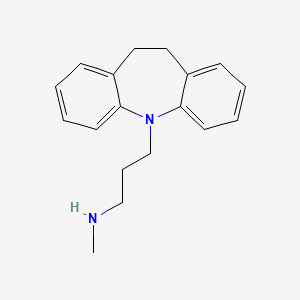 |
0.420 | ||
| ENC001388 | 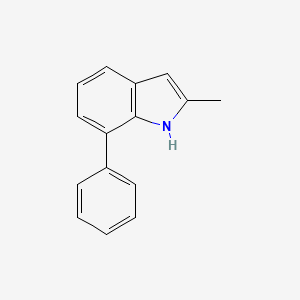 |
0.387 | D04QZD | 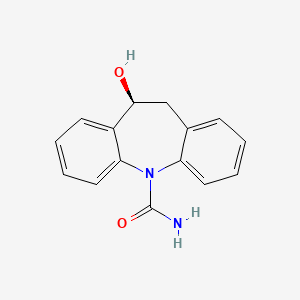 |
0.409 | ||