NPs Basic Information

|
Name |
4-Hydroxybenzoate
|
| Molecular Formula | C7H5O3- | |
| IUPAC Name* |
4-carboxyphenolate
|
|
| SMILES |
C1=CC(=CC=C1C(=O)O)[O-]
|
|
| InChI |
InChI=1S/C7H6O3/c8-6-3-1-5(2-4-6)7(9)10/h1-4,8H,(H,9,10)/p-1
|
|
| InChIKey |
FJKROLUGYXJWQN-UHFFFAOYSA-M
|
|
| Synonyms |
4-hydroxybenzoate; p-hydroxybenzoate; p-Hydroxybenzoate anion; Benzoic acid, 4-hydroxy-, ion(1-); 456-23-5; 4-Hydroxybenzoate ion; 8MJB9HSC8Q; Benzoic acid, p-hydroxy-, ion(1-); 3pcc; 3pch; Hydroxybenzoic acid; 4-hydroxy-benzoate; Benzoic acid, p-hydroxy; Benzoic acid, 4-hydroxy; WLN: QVR DQ; CHEMBL1762656; HSDB 7233; NSC4961; EINECS 202-804-9; DB04242; NCGC00166040-01; AI3-01003; C00156; AE-848/32195059; parahydroxybenzoate; 4-Oxylatobenzoate; 4-oxidanylbenzoate; 4-Carboxyphenoxide; para-hydroxybenzoate; 4e3g; PHB; UNII-8MJB9HSC8Q; H20059_ALDRICH; W398608_ALDRICH; p-Hydroxybenzoic acid monoanion; 240141_ALDRICH; p-Hydroxybenzoic acid ion(1-); 54630_FLUKA; CHEBI:17879; 4-hydroxybenzoic acid, ion(1-); DTXSID10196564; AIDS018038; AIDS-018038; BDBM50340074; P-HYDROXYBENZENECARBOXYLIC ACID; ST5210584; A846111; Q27102693
|
|
| CAS | 456-23-5 | |
| PubChem CID | 54675830 | |
| ChEMBL ID | CHEMBL1762656 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 137.11 | ALogp: | 2.5 |
| HBD: | 1 | HBA: | 3 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 60.4 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.624 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.27 | MDCK Permeability: | 0.00000849 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.003 |
| Human Intestinal Absorption (HIA): | 0.01 | 20% Bioavailability (F20%): | 0.01 |
| 30% Bioavailability (F30%): | 0.308 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.314 | Plasma Protein Binding (PPB): | 38.35% |
| Volume Distribution (VD): | 0.291 | Fu: | 49.50% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.034 | CYP1A2-substrate: | 0.072 |
| CYP2C19-inhibitor: | 0.04 | CYP2C19-substrate: | 0.047 |
| CYP2C9-inhibitor: | 0.033 | CYP2C9-substrate: | 0.178 |
| CYP2D6-inhibitor: | 0.009 | CYP2D6-substrate: | 0.118 |
| CYP3A4-inhibitor: | 0.038 | CYP3A4-substrate: | 0.062 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.575 | Half-life (T1/2): | 0.924 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.048 | Human Hepatotoxicity (H-HT): | 0.449 |
| Drug-inuced Liver Injury (DILI): | 0.794 | AMES Toxicity: | 0.009 |
| Rat Oral Acute Toxicity: | 0.531 | Maximum Recommended Daily Dose: | 0.004 |
| Skin Sensitization: | 0.247 | Carcinogencity: | 0.05 |
| Eye Corrosion: | 0.173 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.37 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000007 |  |
0.636 | D06NVJ | 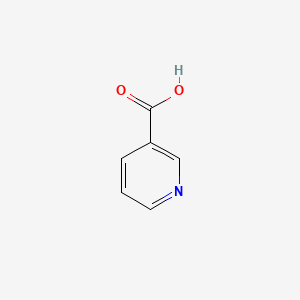 |
0.368 | ||
| ENC000202 |  |
0.553 | D0L7FM |  |
0.368 | ||
| ENC000200 | 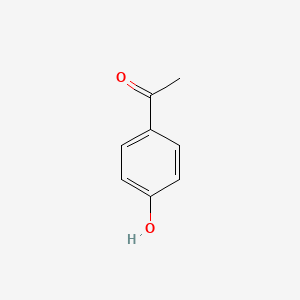 |
0.459 | D0Q8ZX |  |
0.364 | ||
| ENC000665 |  |
0.459 | D07HBX |  |
0.350 | ||
| ENC000013 | 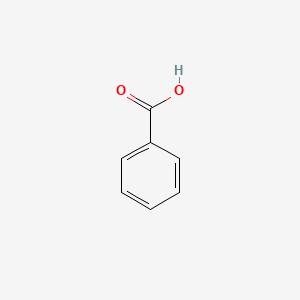 |
0.444 | D06OAV | 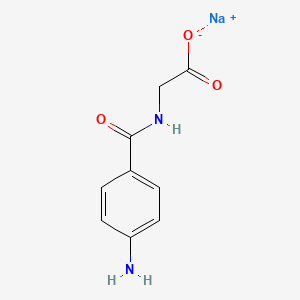 |
0.347 | ||
| ENC000195 | 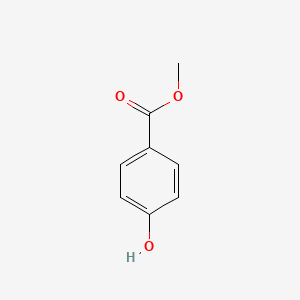 |
0.425 | D0U5QK | 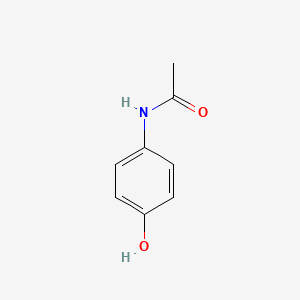 |
0.326 | ||
| ENC002402 |  |
0.421 | D01CRB | 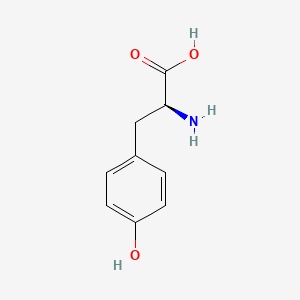 |
0.319 | ||
| ENC005265 |  |
0.420 | D0B3QM | 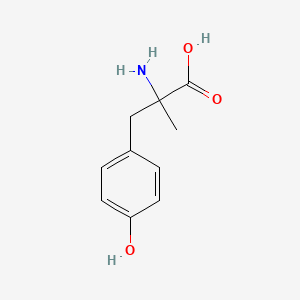 |
0.306 | ||
| ENC005266 |  |
0.420 | D07BPS |  |
0.305 | ||
| ENC003949 |  |
0.420 | D0C4YC | 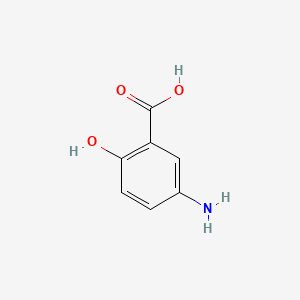 |
0.302 | ||