NPs Basic Information

|
Name |
Radicicol
|
| Molecular Formula | C18H17ClO6 | |
| IUPAC Name* |
(4R,6R,8R,9Z,11E)-16-chloro-17,19-dihydroxy-4-methyl-3,7-dioxatricyclo[13.4.0.06,8]nonadeca-1(15),9,11,16,18-pentaene-2,13-dione
|
|
| SMILES |
C[C@@H]1C[C@@H]2[C@H](O2)/C=C\C=C\C(=O)CC3=C(C(=CC(=C3Cl)O)O)C(=O)O1
|
|
| InChI |
InChI=1S/C18H17ClO6/c1-9-6-15-14(25-15)5-3-2-4-10(20)7-11-16(18(23)24-9)12(21)8-13(22)17(11)19/h2-5,8-9,14-15,21-22H,6-7H2,1H3/b4-2+,5-3-/t9-,14-,15-/m1/s1
|
|
| InChIKey |
WYZWZEOGROVVHK-GTMNPGAYSA-N
|
|
| Synonyms |
radicicol; Monorden; Monorden A; 12772-57-5; (+)-Monorden A; I60EH8GECX; CHEMBL414883; CHEBI:556075; NSC-294404; (1aR,2Z,4E,14R,15aR)-8-chloro-9,11-dihydroxy-14-methyl-1a,14,15,15a-tetrahydro-6H-oxireno[e][2]benzoxacyclotetradecine-6,12(7H)-dione; Monorderne; RDC; Radicolol; Radisicol; RHI-12648; Microlactone, 1; 1bgq; 2wer; 2zbk; 3cgy; 4egk; MFCD06795865; Radicicol R 2146; 2q8i; MONORDEN [MI]; UNII-I60EH8GECX; RADICICOL R-2146; SCHEMBL868832; BDBM15361; BDBM227589; HMS3648C16; 6H-Oxireno(e)(2)benzoxacyclotetradecin-6,12(7H)-dione, 8-chloro-1a,14,15,15a-tetrahydro-9,11-dihydroxy-14-methyl-; HY-N6769; ZINC13521629; AKOS024456686; CCG-208260; DB03758; NSC 294404; NCGC00344093-06; NCGC00344093-09; BR162744; Radicicol from Diheterospora chlamydosporia; CS-0028899; Radicicol from Diheterospora chlamydosporia, solid; SR-01000946352; SR-01000946352-1; Radicicol, Diheterospora chlamydosporia - CAS 12772-57-5; 5-Cl-6-(7,8-epoxy-10-hydroxy-2-oxo-3,5-undecadienyl)-beta-resorcylic acid mu-lactone; (4R,6R,8R,9Z,11E)-16-chloro-17,19-dihydroxy-4-methyl-3,7-dioxatricyclo[13.4.0.0;{6,8}]nonadeca-1(19),9,11,15,17-pentaene-2,13-dione; .BETA.-RESORCYLIC ACID, 5-CHLORO-6-(7,8-EPOXY-10-HYDROXY-2-OXO-3,5-UNDECADIENYL)-, .MU.-LACTONE; RDI
|
|
| CAS | 12772-57-5 | |
| PubChem CID | 6323491 | |
| ChEMBL ID | CHEMBL414883 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 364.8 | ALogp: | 3.4 |
| HBD: | 2 | HBA: | 6 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 96.4 | Aromatic Rings: | 3 |
| Heavy Atoms: | 25 | QED Weighted: | 0.54 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.92 | MDCK Permeability: | 0.00001790 |
| Pgp-inhibitor: | 0.005 | Pgp-substrate: | 0.016 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.013 |
| 30% Bioavailability (F30%): | 0.002 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.131 | Plasma Protein Binding (PPB): | 97.98% |
| Volume Distribution (VD): | 0.603 | Fu: | 1.64% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.91 | CYP1A2-substrate: | 0.091 |
| CYP2C19-inhibitor: | 0.311 | CYP2C19-substrate: | 0.066 |
| CYP2C9-inhibitor: | 0.827 | CYP2C9-substrate: | 0.947 |
| CYP2D6-inhibitor: | 0.77 | CYP2D6-substrate: | 0.289 |
| CYP3A4-inhibitor: | 0.457 | CYP3A4-substrate: | 0.217 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.936 | Half-life (T1/2): | 0.828 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.131 | Human Hepatotoxicity (H-HT): | 0.813 |
| Drug-inuced Liver Injury (DILI): | 0.944 | AMES Toxicity: | 0.391 |
| Rat Oral Acute Toxicity: | 0.741 | Maximum Recommended Daily Dose: | 0.957 |
| Skin Sensitization: | 0.753 | Carcinogencity: | 0.664 |
| Eye Corrosion: | 0.005 | Eye Irritation: | 0.492 |
| Respiratory Toxicity: | 0.902 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC004730 | 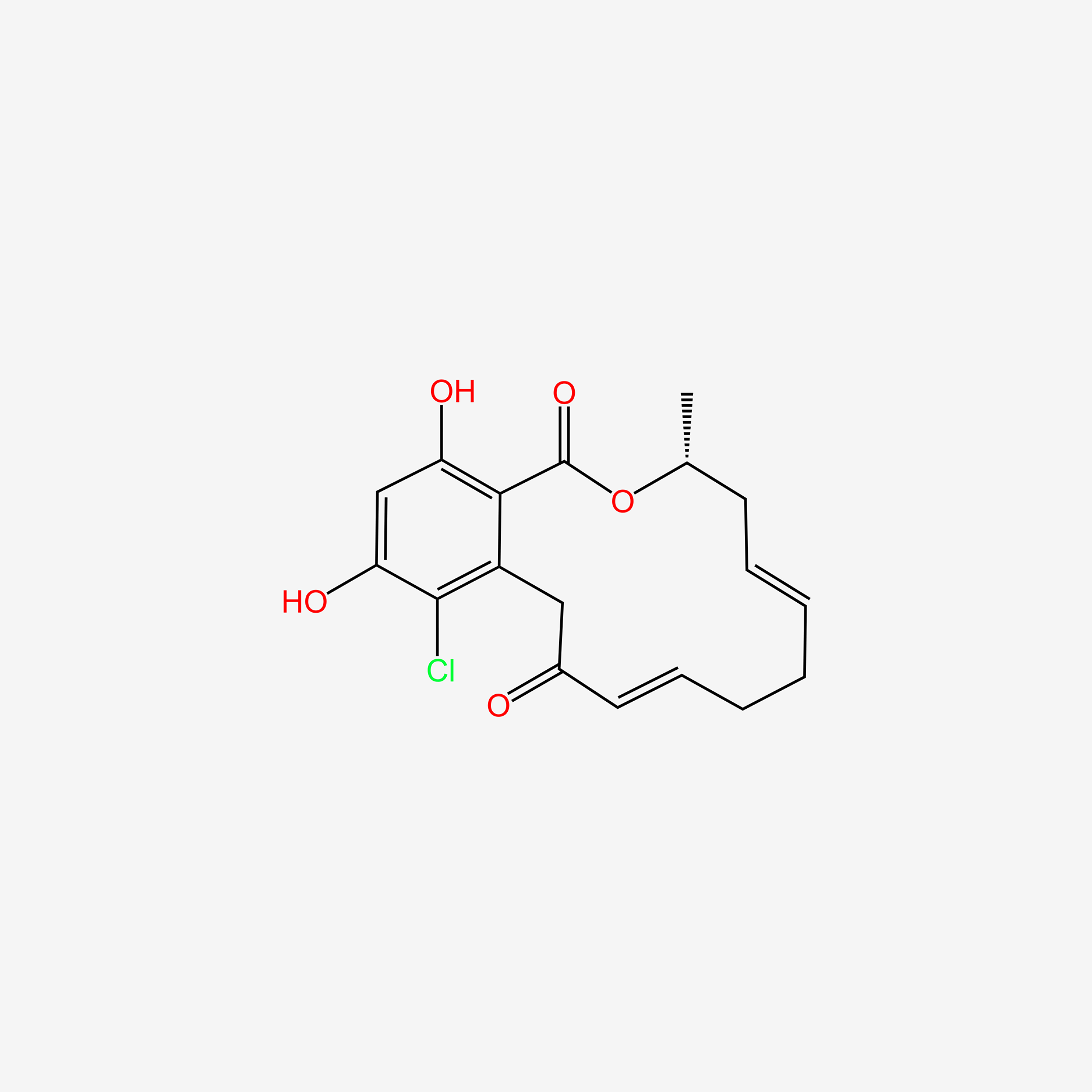 |
0.588 | D07MGA |  |
0.275 | ||
| ENC002592 |  |
0.544 | D0R6BI | 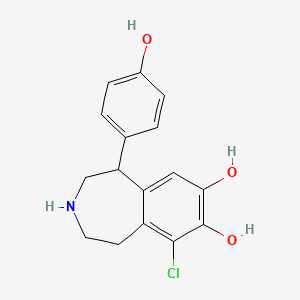 |
0.243 | ||
| ENC002045 |  |
0.493 | D01XDL | 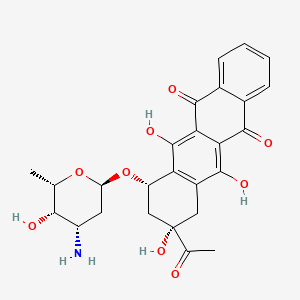 |
0.228 | ||
| ENC002927 | 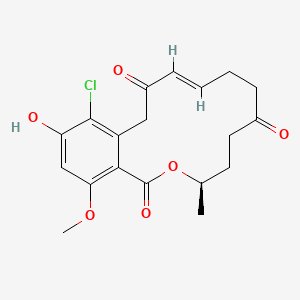 |
0.429 | D0AZ8C |  |
0.219 | ||
| ENC005703 |  |
0.380 | D0C1SF |  |
0.218 | ||
| ENC005418 |  |
0.372 | D0R9WP |  |
0.214 | ||
| ENC003934 |  |
0.366 | D0I9HF |  |
0.214 | ||
| ENC005138 |  |
0.358 | D0H6QU |  |
0.212 | ||
| ENC005939 |  |
0.354 | D01XWG |  |
0.211 | ||
| ENC003117 |  |
0.354 | D0R6RC | 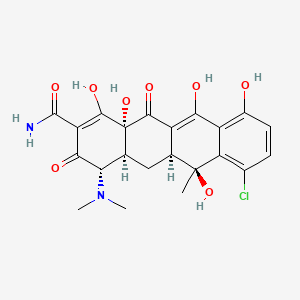 |
0.211 | ||