NPs Basic Information
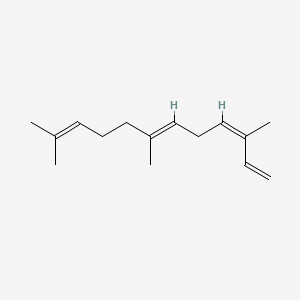
|
Name |
(Z,E)-alpha-Farnesene
|
| Molecular Formula | C15H24 | |
| IUPAC Name* |
(3Z,6E)-3,7,11-trimethyldodeca-1,3,6,10-tetraene
|
|
| SMILES |
CC(=CCC/C(=C/C/C=C(/C)\C=C)/C)C
|
|
| InChI |
InChI=1S/C15H24/c1-6-14(4)10-8-12-15(5)11-7-9-13(2)3/h6,9-10,12H,1,7-8,11H2,2-5H3/b14-10-,15-12+
|
|
| InChIKey |
CXENHBSYCFFKJS-OXYODPPFSA-N
|
|
| Synonyms |
(Z,E)-alpha-Farnesene; 26560-14-5; (3z,6e)-alpha-farnesene; (Z,E)-.alpha.-Farnesene; 1,3,6,10-Dodecatetraene, 3,7,11-trimethyl-, (Z,E)-; cis,trans-.alpha.-Farnesene; 4U4U81627K; (3Z,6E)-3,7,11-trimethyldodeca-1,3,6,10-tetraene; 1,3,6,10-Dodecatetraene, 3,7,11-trimethyl-, (3Z,6E)-; (3Z,6E)-3,7,11-Trimethyl-1,3,6,10-dodecatetraene; cis-.alpha.-Farnesene; trans,trans-alpha-farnesene; cis,trans-alpha-Farnesene; Alph; alpha-Farnesene, (3Z,6E)-; UNII-4U4U81627K; (Z,E)--Farnesene; FEMA No. 3839, (3Z,6E)-alpha-; alpha-trans,trans-Farnesene; .alpha.-(Z,E)-Farnesene; (3Z,6E)- .alpha.-Farnesene; CHEBI:39238; DTXSID70181138; ZINC1531531; (Z)-3, (E)-6-.alpha.-Farnesene; .ALPHA.-FARNESENE, (3Z,6E)-; FEMA NO. 3839, (3Z,6E)-.ALPHA.-; 3,7,11-Trimethyl-(E,E)-1,3,6,10-Dodecatetraene; Q27119780; (Z,E)-3,7,11-Trimethyl-1,3,6,10-dodecatetraene,
|
|
| CAS | 26560-14-5 | |
| PubChem CID | 5362889 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 204.35 | ALogp: | 6.1 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 6 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 15 | QED Weighted: | 0.396 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.475 | MDCK Permeability: | 0.00001900 |
| Pgp-inhibitor: | 0.167 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.213 |
| 30% Bioavailability (F30%): | 0.271 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.757 | Plasma Protein Binding (PPB): | 99.56% |
| Volume Distribution (VD): | 3.948 | Fu: | 1.94% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.951 | CYP1A2-substrate: | 0.262 |
| CYP2C19-inhibitor: | 0.67 | CYP2C19-substrate: | 0.779 |
| CYP2C9-inhibitor: | 0.42 | CYP2C9-substrate: | 0.329 |
| CYP2D6-inhibitor: | 0.473 | CYP2D6-substrate: | 0.126 |
| CYP3A4-inhibitor: | 0.439 | CYP3A4-substrate: | 0.272 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.51 | Half-life (T1/2): | 0.299 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.008 | Human Hepatotoxicity (H-HT): | 0.96 |
| Drug-inuced Liver Injury (DILI): | 0.027 | AMES Toxicity: | 0.059 |
| Rat Oral Acute Toxicity: | 0.01 | Maximum Recommended Daily Dose: | 0.876 |
| Skin Sensitization: | 0.941 | Carcinogencity: | 0.267 |
| Eye Corrosion: | 0.952 | Eye Irritation: | 0.984 |
| Respiratory Toxicity: | 0.868 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001564 | 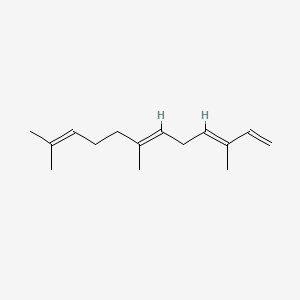 |
1.000 | D05XQE |  |
0.429 | ||
| ENC001566 |  |
0.729 | D09XWD | 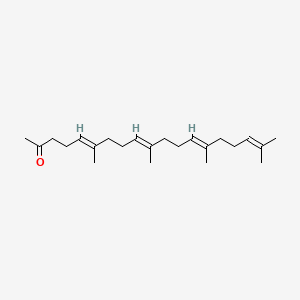 |
0.400 | ||
| ENC000526 | 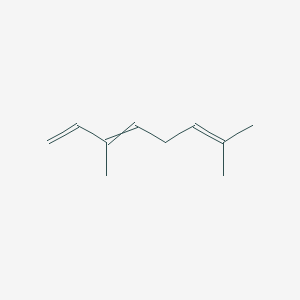 |
0.610 | D03VFL | 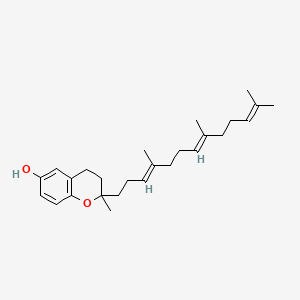 |
0.326 | ||
| ENC001568 | 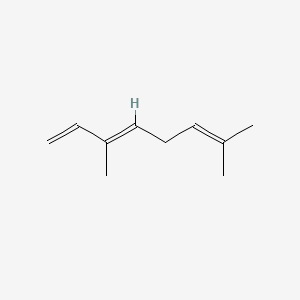 |
0.610 | D0M1PQ | 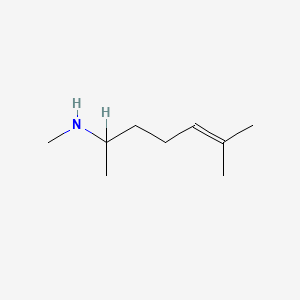 |
0.294 | ||
| ENC001467 | 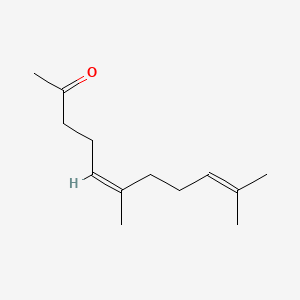 |
0.540 | D06BLQ | 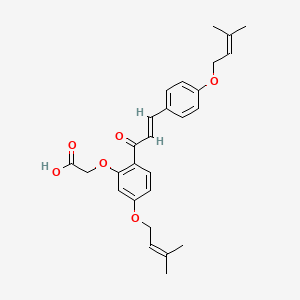 |
0.200 | ||
| ENC001717 | 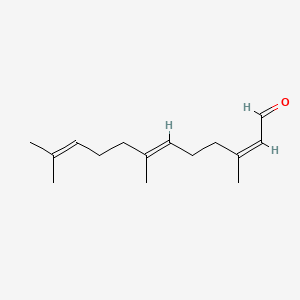 |
0.537 | D0H6VY |  |
0.188 | ||
| ENC001096 | 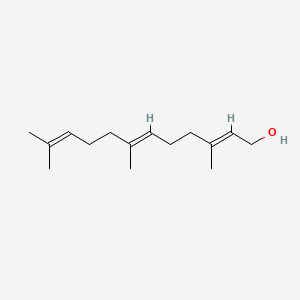 |
0.537 | D03ZFG | 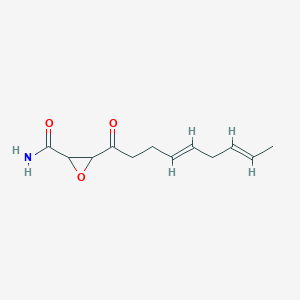 |
0.183 | ||
| ENC001462 | 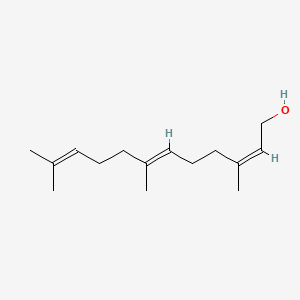 |
0.537 | D0S7WX | 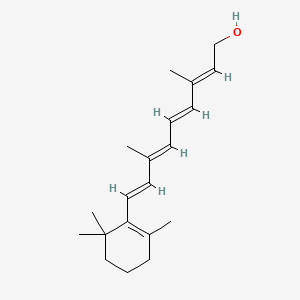 |
0.183 | ||
| ENC002413 | 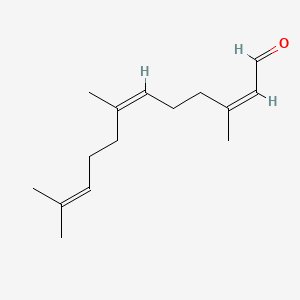 |
0.537 | D0X7XG | 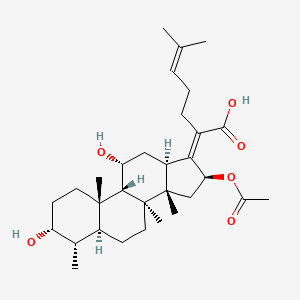 |
0.168 | ||
| ENC000314 | 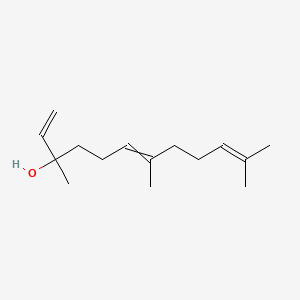 |
0.519 | D02DGU | 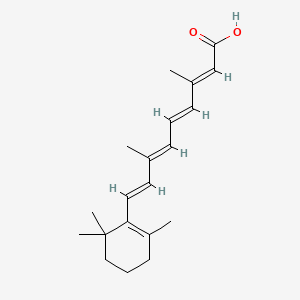 |
0.165 | ||