NPs Basic Information
|
Name |
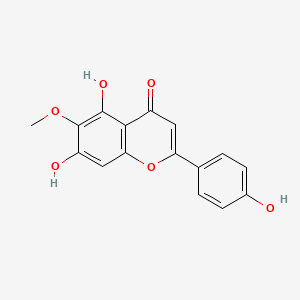
Hispidulin
|
| Molecular Formula | C16H12O6 | |
| IUPAC Name* |
5,7-dihydroxy-2-(4-hydroxyphenyl)-6-methoxychromen-4-one
|
|
| SMILES |
COC1=C(C2=C(C=C1O)OC(=CC2=O)C3=CC=C(C=C3)O)O
|
|
| InChI |
InChI=1S/C16H12O6/c1-21-16-11(19)7-13-14(15(16)20)10(18)6-12(22-13)8-2-4-9(17)5-3-8/h2-7,17,19-20H,1H3
|
|
| InChIKey |
IHFBPDAQLQOCBX-UHFFFAOYSA-N
|
|
| Synonyms |
Hispidulin; 1447-88-7; Dinatin; Scutellarein 6-methyl ether; 4',5,7-Trihydroxy-6-methoxyflavone; 4H-1-BENZOPYRAN-4-ONE, 5,7-DIHYDROXY-2-(4-HYDROXYPHENYL)-6-METHOXY-; 6-O-Methylapigenin; Salvitin; 5,7-dihydroxy-2-(4-hydroxyphenyl)-6-methoxychromen-4-one; TCMDC-123942; 5,7-Dihydroxy-2-(4-hydroxyphenyl)-6-methoxy-4H-chromen-4-one; NSC 122415; NSC-122415; NSC122415; Flavone, 4',5,7-trihydroxy-6-methoxy-; CHEMBL293776; CHEBI:75902; N7F61604C2; 5,7-Dihydroxy-2-(4-hydroxyphenyl)-6-methoxy-4H-1-benzopyran-4-one; methoxyapigenin; 6-methoxyapigenin; CCRIS 8484; 6-methoxy apigenin; HISPEDULIN; M-3-GHYDRATE; Oprea1_873387; MLS000728540; SCHEMBL514926; MEGxp0_000683; Hispidulin, >=98% (HPLC); UNII-N7F61604C2; ACon1_000933; cid_5281628; DTXSID30162786; PubChem SID: 26725244; 4',7-Trihydroxy-6-methoxyflavone; 5,7-dihydroxy-2-(4-hydroxyphenyl)-6-methoxy-chromen-4-one; HMS2223A03; HMS3344G13; HMS3868N13; HY-N1950; ZINC5732241; Flavone,5,7-trihydroxy-6-methoxy-; BDBM50049395; LMPK12111159; MFCD00143504; s3296; ZB1763; 4?,5,7-Trihydroxy-6-methoxyflavone; AKOS004110694; 5,7,4''-Trihydroxy-6-methoxyflavone; CS-6502; DB14008; NCGC00167728-01; NCGC00167728-02; NCGC00169216-01; AC-34245; AS-78830; NCI60_000530; SMR000445653; FT-0697687; 447H887; Q-100165; BRD-K72066874-001-01-0; Q15410994; 5,7-Dihydroxy-2-(4-hydroxy-phenyl)-6-methoxy-chromen-4-one; 4H-1-Benzopyran-4-one,7-dihydroxy-2-(4-hydroxyphenyl)-6-methoxy-; 5,7-Dihydroxy-2-(4-hydroxyphenyl)-6-methoxy-4H-chromen-4-one #; 4H-1-Benzopyran-4-one, 5, 7-dihydroxy-2-(4-hydroxyphenyl)-6-methoxy-; HUL
|
|
| CAS | 1447-88-7 | |
| PubChem CID | 5281628 | |
| ChEMBL ID | CHEMBL293776 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 300.26 | ALogp: | 1.7 |
| HBD: | 3 | HBA: | 6 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 96.2 | Aromatic Rings: | 3 |
| Heavy Atoms: | 22 | QED Weighted: | 0.671 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.882 | MDCK Permeability: | 0.00001230 |
| Pgp-inhibitor: | 0.011 | Pgp-substrate: | 0.928 |
| Human Intestinal Absorption (HIA): | 0.017 | 20% Bioavailability (F20%): | 0.463 |
| 30% Bioavailability (F30%): | 0.985 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.004 | Plasma Protein Binding (PPB): | 96.96% |
| Volume Distribution (VD): | 0.52 | Fu: | 7.81% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.971 | CYP1A2-substrate: | 0.824 |
| CYP2C19-inhibitor: | 0.391 | CYP2C19-substrate: | 0.059 |
| CYP2C9-inhibitor: | 0.686 | CYP2C9-substrate: | 0.894 |
| CYP2D6-inhibitor: | 0.676 | CYP2D6-substrate: | 0.495 |
| CYP3A4-inhibitor: | 0.524 | CYP3A4-substrate: | 0.156 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.798 | Half-life (T1/2): | 0.857 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.099 | Human Hepatotoxicity (H-HT): | 0.057 |
| Drug-inuced Liver Injury (DILI): | 0.913 | AMES Toxicity: | 0.395 |
| Rat Oral Acute Toxicity: | 0.073 | Maximum Recommended Daily Dose: | 0.139 |
| Skin Sensitization: | 0.906 | Carcinogencity: | 0.225 |
| Eye Corrosion: | 0.004 | Eye Irritation: | 0.924 |
| Respiratory Toxicity: | 0.241 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001533 | 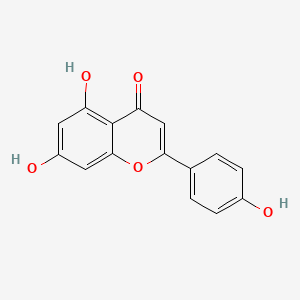 |
0.696 | D06GCK |  |
0.625 | ||
| ENC001751 | 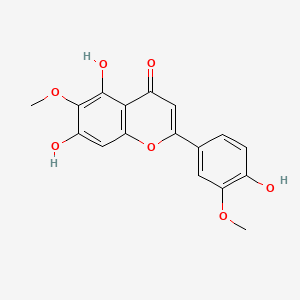 |
0.693 | D04AIT |  |
0.545 | ||
| ENC001548 |  |
0.566 | D0K8KX | 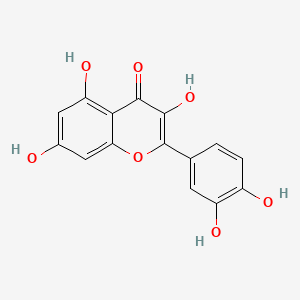 |
0.424 | ||
| ENC001534 | 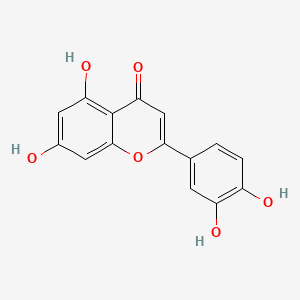 |
0.545 | D07MGA |  |
0.356 | ||
| ENC002757 | 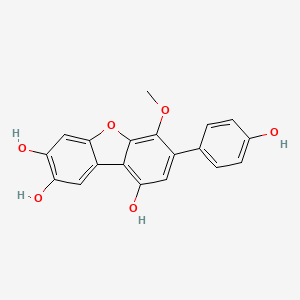 |
0.541 | D0R6BI | 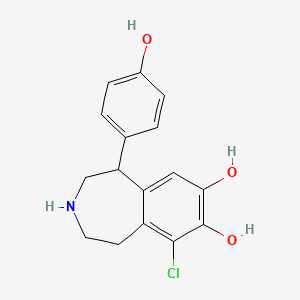 |
0.319 | ||
| ENC002475 | 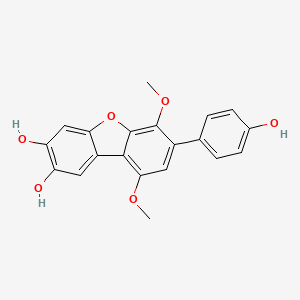 |
0.523 | D08SKH | 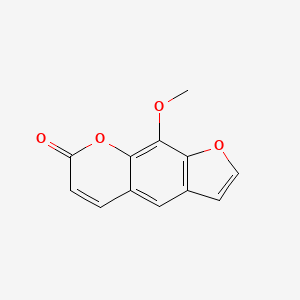 |
0.305 | ||
| ENC001550 |  |
0.500 | D06TJJ | 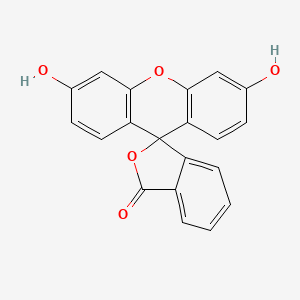 |
0.304 | ||
| ENC001771 |  |
0.500 | D04XEG | 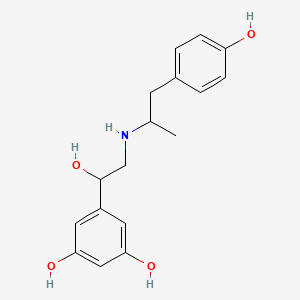 |
0.298 | ||
| ENC003492 | 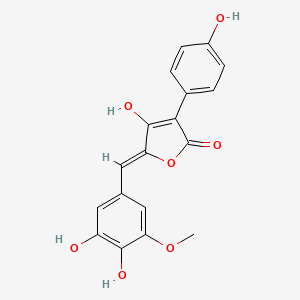 |
0.494 | D0TC7C | 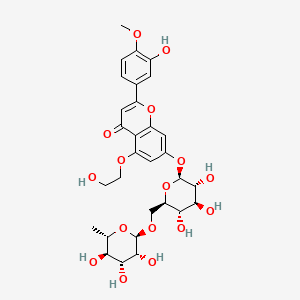 |
0.297 | ||
| ENC002625 | 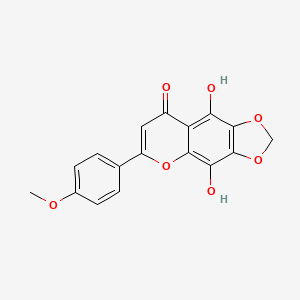 |
0.483 | D0G4KG | 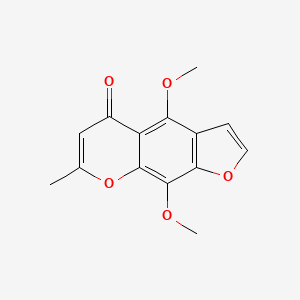 |
0.295 | ||