NPs Basic Information
|
Name |
Palmitoleic Acid
|
| Molecular Formula | C16H30O2 | |
| IUPAC Name* |
(Z)-hexadec-9-enoic acid
|
|
| SMILES |
CCCCCC/C=C\CCCCCCCC(=O)O
|
|
| InChI |
InChI=1S/C16H30O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16(17)18/h7-8H,2-6,9-15H2,1H3,(H,17,18)/b8-7-
|
|
| InChIKey |
SECPZKHBENQXJG-FPLPWBNLSA-N
|
|
| Synonyms |
palmitoleic acid; 373-49-9; (Z)-Hexadec-9-enoic acid; cis-9-Hexadecenoic acid; 9-cis-Hexadecenoic acid; (Z)-9-hexadecenoic acid; zoomaric acid; Zoomeric acid; cis-Palmitoleic acid; Oleopalmitic acid; cis-9-palmitoleic acid; (9Z)-hexadec-9-enoic acid; (9Z)-Hexadecenoic acid; palmitoleate; Palmitolinoleic acid; 9Z-hexadecenoic acid; 9Z-palmitoleic acid; Hexadecenoate; Oleopalmitate; Zoomerate; cis-Palmitoleate; 9-Hexadecenoate; 9-cis-hexadecenoate; cis-delta-9-Hexadecenoic acid; cis-Delta(9)-hexadecenoic acid; (Z)-9-hexadecenoate; (Z)-Palmitoleic acid; 9-Hexadecenoic acid, (9Z)-; 16:1Delta9; cis-9-hexadecenoate; cis-delta-9-Hexadecenoate; (Z)-Hexadec-9-enoicacid; 9-Hexadecenoic acid, (Z)-; (Z)-hexadec-9-enoate; 209B6YPZ4I; CHEMBL453509; CHEBI:28716; C16:1n-7; FA 16:1; cis-.delta.9-Hexadecenoic acid; UNII-209B6YPZ4I; palmitoleic-acid; Physetoleic acid; NSC277452; NSC-277452; cis-Palmitoleicacid; EINECS 206-765-9; MFCD00004437; NSC 277452; AI3-36443; (Z)-9-hexadecenic acid; bmse000926; DSSTox_CID_21197; DSSTox_RID_79644; Hexadecenoate (n-C16:1); DSSTox_GSID_41197; SCHEMBL33310; GTPL5547; DTXSID0041197; PALMITOLEIC ACID [WHO-DD]; ZINC8221009; Tox21_300950; BDBM50269531; LMFA01030056; Palmitoleic acid, analytical standard; AKOS015843187; CS-W012589; DB04257; FS-6301; HY-W011873; NCGC00248225-01; NCGC00248225-03; NCGC00254852-01; AC-33786; CAS-373-49-9; FA(16:1(9Z)); H0072; Palmitoleic acid, >=98.5% (GC), liquid; S3341; C08362; Q412366; 16:1(N-7); ED9A824A-5CB7-476E-884E-4216DD38386E; PALMITOLEIC ACID (CONSTITUENT OF SPIRULINA) [DSC]; PALMITOLEIC ACID (CONSTITUENT OF SAW PALMETTO) [DSC]
|
|
| CAS | 373-49-9 | |
| PubChem CID | 445638 | |
| ChEMBL ID | CHEMBL453509 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 254.41 | ALogp: | 6.4 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 13 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 37.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 18 | QED Weighted: | 0.344 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.008 | MDCK Permeability: | 0.00003800 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.03 | 20% Bioavailability (F20%): | 0.99 |
| 30% Bioavailability (F30%): | 0.997 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.043 | Plasma Protein Binding (PPB): | 98.32% |
| Volume Distribution (VD): | 0.543 | Fu: | 0.73% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.255 | CYP1A2-substrate: | 0.269 |
| CYP2C19-inhibitor: | 0.11 | CYP2C19-substrate: | 0.185 |
| CYP2C9-inhibitor: | 0.271 | CYP2C9-substrate: | 0.988 |
| CYP2D6-inhibitor: | 0.013 | CYP2D6-substrate: | 0.293 |
| CYP3A4-inhibitor: | 0.037 | CYP3A4-substrate: | 0.029 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.631 | Half-life (T1/2): | 0.84 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.043 | Human Hepatotoxicity (H-HT): | 0.054 |
| Drug-inuced Liver Injury (DILI): | 0.017 | AMES Toxicity: | 0.017 |
| Rat Oral Acute Toxicity: | 0.044 | Maximum Recommended Daily Dose: | 0.028 |
| Skin Sensitization: | 0.921 | Carcinogencity: | 0.12 |
| Eye Corrosion: | 0.937 | Eye Irritation: | 0.97 |
| Respiratory Toxicity: | 0.758 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
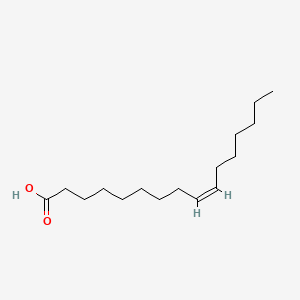
| ENC001589 | 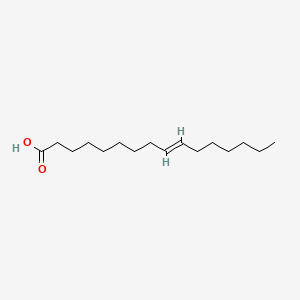 |
1.000 | D0O1PH |  |
0.773 | ||
| ENC001555 | 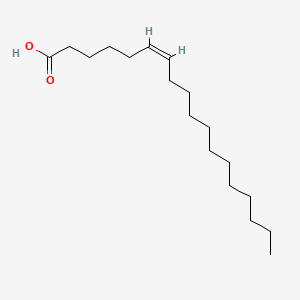 |
0.895 | D0O1TC | 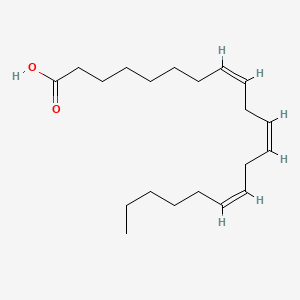 |
0.701 | ||
| ENC001591 | 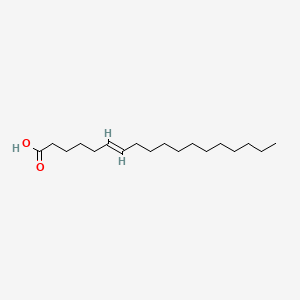 |
0.895 | D0UE9X | 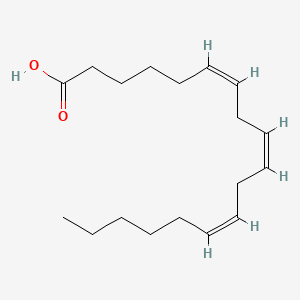 |
0.612 | ||
| ENC001419 | 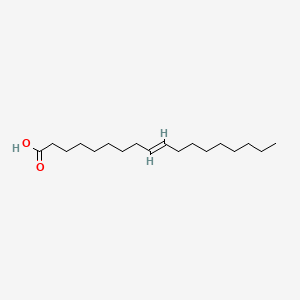 |
0.895 | D0Z5BC | 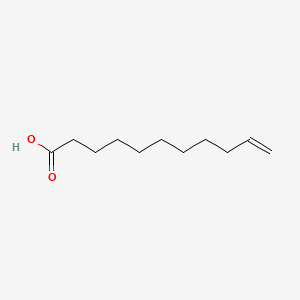 |
0.554 | ||
| ENC001592 | 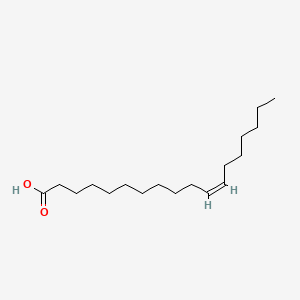 |
0.895 | D0OR6A | 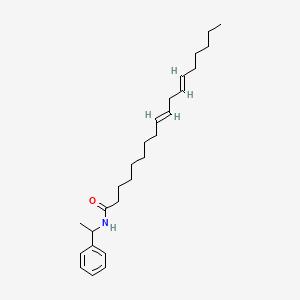 |
0.500 | ||
| ENC001100 | 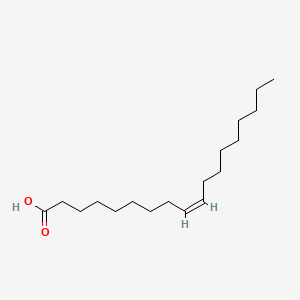 |
0.895 | D0XN8C | 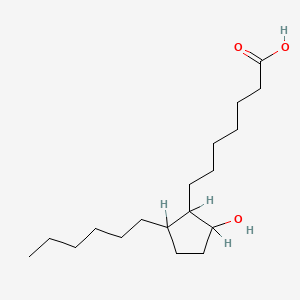 |
0.486 | ||
| ENC001554 |  |
0.846 | D09SRR |  |
0.482 | ||
| ENC001544 |  |
0.831 | D07ILQ |  |
0.461 | ||
| ENC001535 |  |
0.831 | D05ATI |  |
0.420 | ||
| ENC001584 |  |
0.831 | D0I4DQ |  |
0.412 | ||