NPs Basic Information
|
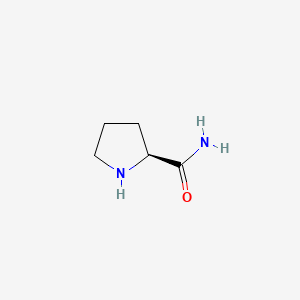
Name |
L-prolinamide
|
| Molecular Formula | C5H10N2O | |
| IUPAC Name* |
(2S)-pyrrolidine-2-carboxamide
|
|
| SMILES |
C1C[C@H](NC1)C(=O)N
|
|
| InChI |
InChI=1S/C5H10N2O/c6-5(8)4-2-1-3-7-4/h4,7H,1-3H2,(H2,6,8)/t4-/m0/s1
|
|
| InChIKey |
VLJNHYLEOZPXFW-BYPYZUCNSA-N
|
|
| Synonyms |
L-prolinamide; 7531-52-4; Prolinamide; h-pro-nh2; (S)-Pyrrolidine-2-carboxamide; (S)-Prolinamide; (2S)-pyrrolidine-2-carboxamide; L-proline amide; l-Prolineamide; (S)-2-Pyrrolidinecarboxamide; 58274-20-7; proline amide; (S)-proline amide; 2-pyrrolidinecarboxamide,(s)-; l-(-)-prolinamide; VD6PQK9DHG; (2S)-2-pyrrolidinecarboxamide; CHEBI:21374; MFCD00005253; (s)-pyrrolidine-2-carboxylic acid amide; 2-PYRROLIDINECARBOXAMIDE, (2S)-; UNII-VD6PQK9DHG; (2S)-2-Carbamoylpyrrolidine; proline imide; (2S)pyrrolidine-2-carboxamide; L-proline amid; (2S)-prolinamide; LPD; EINECS 231-397-0; ProNH2; L-(-) prolinamide; H-ProNH2; L-ProNH2; L-Prolinamide, 98%; PROLINAMIDE, L-; 2-Pyrrolidinecarboxamide #; EC 231-397-0; H-L-PRO-NH2; SCHEMBL240170; (S)-Prrolidine-2-carboxamide; (S)-2-pyrrolidine carboxamide; CHEMBL1222059; 2-Pyrrolidinecarboxamide, (S)-; DTXSID00226268; ZINC391898; ACT05030; ALBB-014823; CS-D1209; AKOS005174571; AKOS015854494; NCGC00159402-02; AC-24602; AC-25877; BP-10428; HY-20582; TS-01579; DB-029980; AM20100732; P1382; A26952; C19781; EN300-114103; S)-PYRROLIDINE-2-CARBOXYLIC ACID AMIDE; (2S)-Pyrrolidine-2-carboxamide (L-Prolinamide); 429P276; A839595; J-524162; Q-102892; Q27103084; F1905-7145; (S)-Pyrrolidine-2-carboxylic acid amide;(S)-Prolinamide; L-Proline amide;L-(-)-Prolinamide
|
|
| CAS | 7531-52-4 | |
| PubChem CID | 111306 | |
| ChEMBL ID | CHEMBL1222059 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Physi-Chem Properties
| Molecular Weight: | 114.15 | ALogp: | -0.9 |
| HBD: | 2 | HBA: | 2 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 55.1 | Aromatic Rings: | 1 |
| Heavy Atoms: | 8 | QED Weighted: | 0.491 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.18 | MDCK Permeability: | 0.00013336 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.091 |
| Human Intestinal Absorption (HIA): | 0.023 | 20% Bioavailability (F20%): | 0.005 |
| 30% Bioavailability (F30%): | 0.101 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.168 | Plasma Protein Binding (PPB): | 10.91% |
| Volume Distribution (VD): | 1.268 | Fu: | 92.80% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.018 | CYP1A2-substrate: | 0.084 |
| CYP2C19-inhibitor: | 0.027 | CYP2C19-substrate: | 0.14 |
| CYP2C9-inhibitor: | 0.004 | CYP2C9-substrate: | 0.123 |
| CYP2D6-inhibitor: | 0.004 | CYP2D6-substrate: | 0.407 |
| CYP3A4-inhibitor: | 0.005 | CYP3A4-substrate: | 0.145 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.807 | Half-life (T1/2): | 0.413 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.022 | Human Hepatotoxicity (H-HT): | 0.234 |
| Drug-inuced Liver Injury (DILI): | 0.059 | AMES Toxicity: | 0.046 |
| Rat Oral Acute Toxicity: | 0.194 | Maximum Recommended Daily Dose: | 0.08 |
| Skin Sensitization: | 0.353 | Carcinogencity: | 0.383 |
| Eye Corrosion: | 0.017 | Eye Irritation: | 0.109 |
| Respiratory Toxicity: | 0.253 |