NPs Basic Information
|
Name |
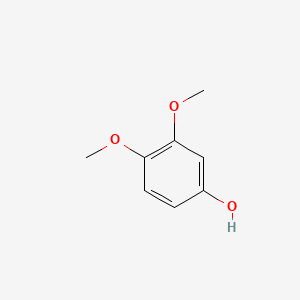
3,4-Dimethoxyphenol
|
| Molecular Formula | C8H10O3 | |
| IUPAC Name* |
3,4-dimethoxyphenol
|
|
| SMILES |
COC1=C(C=C(C=C1)O)OC
|
|
| InChI |
InChI=1S/C8H10O3/c1-10-7-4-3-6(9)5-8(7)11-2/h3-5,9H,1-2H3
|
|
| InChIKey |
SMFFZOQLHYIRDA-UHFFFAOYSA-N
|
|
| Synonyms |
3,4-DIMETHOXYPHENOL; 2033-89-8; Phenol, 3,4-dimethoxy-; 3,4-Bis(methyloxy)phenol; 3,4-dimethoxy phenol; 38B43WCU83; MFCD00008390; 4-Hydroxyveratrole; NSC-140927; 3,4-dimethoxyphel; UNII-38B43WCU83; EINECS 217-995-4; NSC140927; 3,4,dimethoxyphenol; Phenol,4-dimethoxy-; NSC 140927; 3,4-dimethoxy-phenol; 3, 4-dimethoxy phenol; 3,4-Dimethoxyphenol, 97%; SCHEMBL119396; 1-hydroxy-3,4-dimethoxybenzene; DTXSID7062112; ZINC388573; ACT11670; CS-B1823; HY-N1780; STR06225; AKOS005146109; AC-3793; AM61832; SY017636; TS-02054; DB-031011; A4425; D3221; FT-0614348; EN300-91592; 033D898; AE-508/42302277; Q-101910; Q27256773
|
|
| CAS | 2033-89-8 | |
| PubChem CID | 16251 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 154.16 | ALogp: | 1.7 |
| HBD: | 1 | HBA: | 3 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 38.7 | Aromatic Rings: | 1 |
| Heavy Atoms: | 11 | QED Weighted: | 0.707 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.463 | MDCK Permeability: | 0.00001930 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.515 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.015 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.337 | Plasma Protein Binding (PPB): | 59.51% |
| Volume Distribution (VD): | 1.216 | Fu: | 26.46% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.839 | CYP1A2-substrate: | 0.917 |
| CYP2C19-inhibitor: | 0.285 | CYP2C19-substrate: | 0.842 |
| CYP2C9-inhibitor: | 0.037 | CYP2C9-substrate: | 0.899 |
| CYP2D6-inhibitor: | 0.122 | CYP2D6-substrate: | 0.918 |
| CYP3A4-inhibitor: | 0.071 | CYP3A4-substrate: | 0.281 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.846 | Half-life (T1/2): | 0.905 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.048 | Human Hepatotoxicity (H-HT): | 0.075 |
| Drug-inuced Liver Injury (DILI): | 0.197 | AMES Toxicity: | 0.026 |
| Rat Oral Acute Toxicity: | 0.091 | Maximum Recommended Daily Dose: | 0.072 |
| Skin Sensitization: | 0.778 | Carcinogencity: | 0.287 |
| Eye Corrosion: | 0.914 | Eye Irritation: | 0.978 |
| Respiratory Toxicity: | 0.379 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000712 | 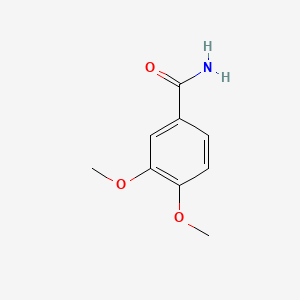 |
0.585 | D0E9CD | 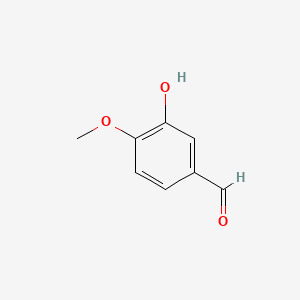 |
0.429 | ||
| ENC000478 | 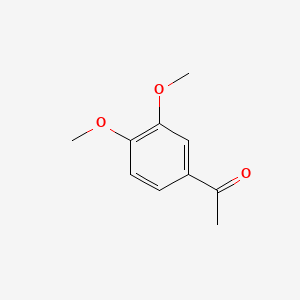 |
0.585 | D09GYT | 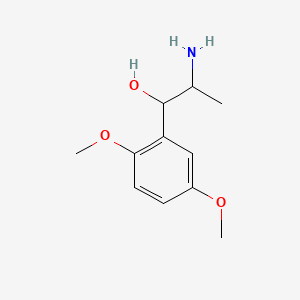 |
0.429 | ||
| ENC000168 | 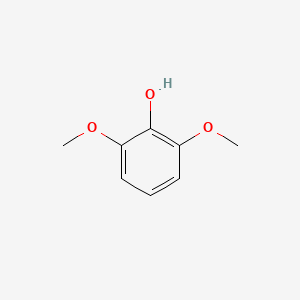 |
0.579 | D0Q9ON | 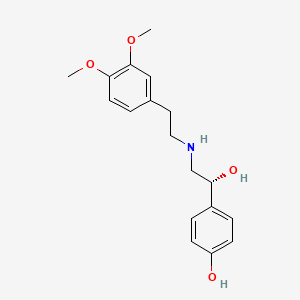 |
0.418 | ||
| ENC001461 | 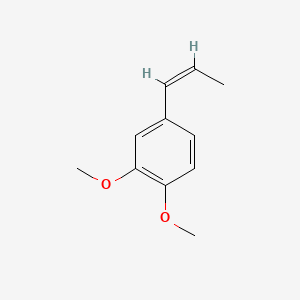 |
0.571 | D02XJY | 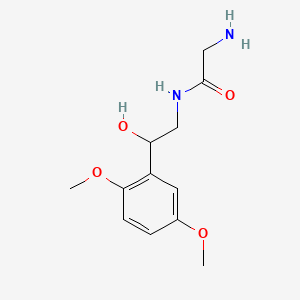 |
0.362 | ||
| ENC000499 | 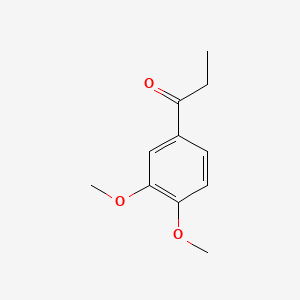 |
0.545 | D06GCK |  |
0.356 | ||
| ENC001363 | 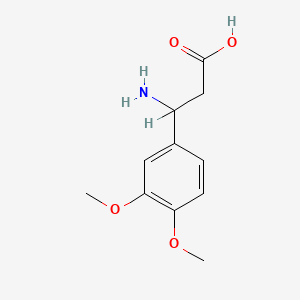 |
0.521 | D0E6OC | 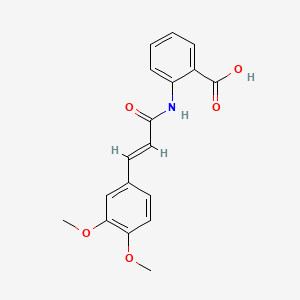 |
0.347 | ||
| ENC002891 | 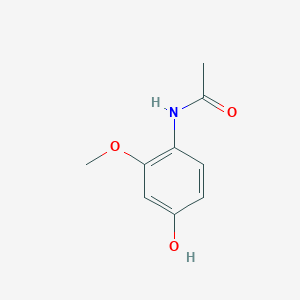 |
0.512 | D01SAT | 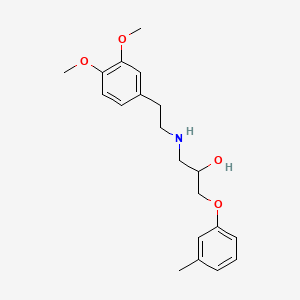 |
0.329 | ||
| ENC000172 |  |
0.462 | D0NJ3V | 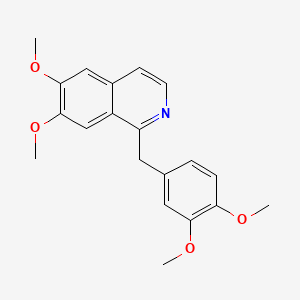 |
0.312 | ||
| ENC005746 |  |
0.451 | D0S5LH | 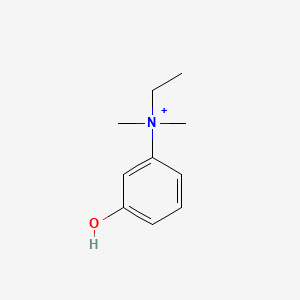 |
0.292 | ||
| ENC000304 | 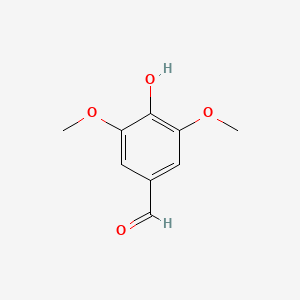 |
0.444 | D0F4ZY |  |
0.289 | ||