NPs Basic Information

|
Name |
2,3,5,6-Tetramethylpyrazine
|
| Molecular Formula | C8H12N2 | |
| IUPAC Name* |
2,3,5,6-tetramethylpyrazine
|
|
| SMILES |
CC1=C(N=C(C(=N1)C)C)C
|
|
| InChI |
InChI=1S/C8H12N2/c1-5-6(2)10-8(4)7(3)9-5/h1-4H3
|
|
| InChIKey |
FINHMKGKINIASC-UHFFFAOYSA-N
|
|
| Synonyms |
2,3,5,6-Tetramethylpyrazine; TETRAMETHYLPYRAZINE; 1124-11-4; Ligustrazine; Pyrazine, tetramethyl-; Bs factor; Tetrapyrazine; Tetramethylpyrazin; Chuanxiongzine; Liqustrazine; Ligustizine; 2,3,5,6-Tetramethyl pyrazine; chuanxingzine; FEMA No. 3237; 2,3,5,6,-Tetramethyl-1,4-pyrazine; MFCD00006146; V80F4IA5XG; MLS000069594; FEMA 3237; Pyrazine, 2,3,5,6-tetramethyl-; NSC-36080; NSC-46451; SMR000059042; 2,5,6-Tetramethylpyrazine; TMPZ; EINECS 214-391-2; NSC 36080; NSC 46451; UNII-V80F4IA5XG; 2,3,5,6-Tetramethylpyrazine (natural); Ligustrazin; CHUANXIONGQIN; Opera_ID_849; TMP?; DSSTox_CID_27070; DSSTox_RID_82085; DSSTox_GSID_47070; SCHEMBL77624; LIGUSTRAZINE [WHO-DD]; CHEMBL303697; ZINC4042; 2,3,5,6-Tetramethyl-pyrazine; DTXSID6047070; FINHMKGKINIASC-UHFFFAOYSA-; CHEBI:133246; Pyrazine, 2,3,5,6-tetramethyl; HMS2235K03; HMS3371J08; Nat.2,3,5,6-Tetramethylpyrazine; HY-N0264; NSC36080; NSC46451; Tox21_302313; BBL012277; s3956; STL163591; 2,3,5,6-Tetramethylpyrazine, 98%; AKOS003398567; CCG-207974; CS-W023183; NCGC00247063-01; NCGC00256097-01; AC-10515; AC-34076; AS-13206; SY011353; CAS-1124-11-4; DB-003786; 2,3,5,6-TETRAMETHYLPYRAZINE [FCC]; AM20070299; FT-0609443; T0972; 2,3,5,6-TETRAMETHYLPYRAZINE [FHFI]; 2,3,5,6-Tetramethylpyrazine, >=98%, FG; EN300-113945; F11202; 2,3,5,6-Tetramethylpyrazine, analytical standard; A802574; AC-907/25014219; Q-100069; 2,3,5,6-Tetramethylpyrazine, natural, >=98%, FG; Q11319317; Z1741976694; 2,3,5,6-Tetramethylpyrazine, Vetec(TM) reagent grade, 98%
|
|
| CAS | 1124-11-4 | |
| PubChem CID | 14296 | |
| ChEMBL ID | CHEMBL303697 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 136.19 | ALogp: | 1.3 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 25.8 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.547 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.845 | MDCK Permeability: | 0.00002420 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.007 |
| Human Intestinal Absorption (HIA): | 0.011 | 20% Bioavailability (F20%): | 0.005 |
| 30% Bioavailability (F30%): | 0.007 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.975 | Plasma Protein Binding (PPB): | 60.36% |
| Volume Distribution (VD): | 1.392 | Fu: | 47.09% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.386 | CYP1A2-substrate: | 0.905 |
| CYP2C19-inhibitor: | 0.018 | CYP2C19-substrate: | 0.497 |
| CYP2C9-inhibitor: | 0.001 | CYP2C9-substrate: | 0.087 |
| CYP2D6-inhibitor: | 0.151 | CYP2D6-substrate: | 0.846 |
| CYP3A4-inhibitor: | 0.013 | CYP3A4-substrate: | 0.558 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.038 | Half-life (T1/2): | 0.266 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.014 | Human Hepatotoxicity (H-HT): | 0.203 |
| Drug-inuced Liver Injury (DILI): | 0.244 | AMES Toxicity: | 0.126 |
| Rat Oral Acute Toxicity: | 0.436 | Maximum Recommended Daily Dose: | 0.015 |
| Skin Sensitization: | 0.296 | Carcinogencity: | 0.883 |
| Eye Corrosion: | 0.928 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.56 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000646 |  |
0.343 | D02LPF |  |
0.204 | ||
| ENC000909 |  |
0.308 | D0V9YR | 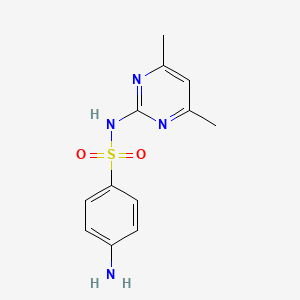 |
0.182 | ||
| ENC000342 | 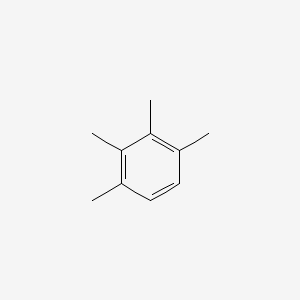 |
0.300 | D0U2CV |  |
0.172 | ||
| ENC000181 | 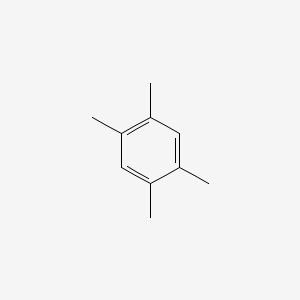 |
0.300 | D0U3DU | 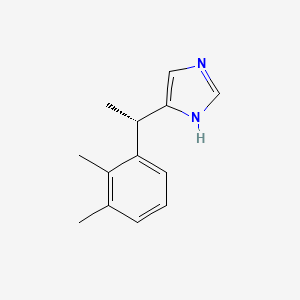 |
0.172 | ||
| ENC001337 |  |
0.286 | D0Y4DY | 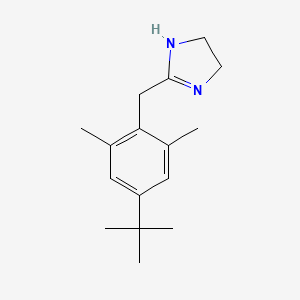 |
0.172 | ||
| ENC001374 |  |
0.240 | D06PQT | 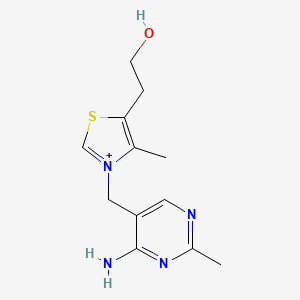 |
0.169 | ||
| ENC000728 | 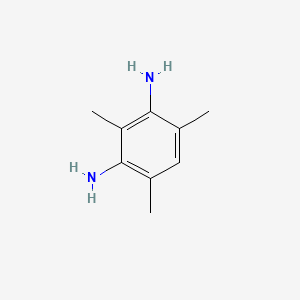 |
0.227 | D09EBS | 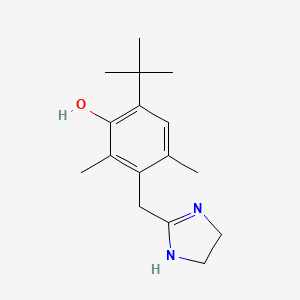 |
0.167 | ||
| ENC002336 | 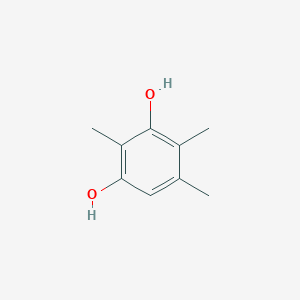 |
0.227 | D02LDV | 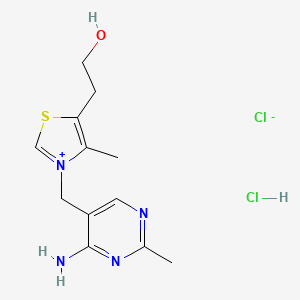 |
0.164 | ||
| ENC005230 | 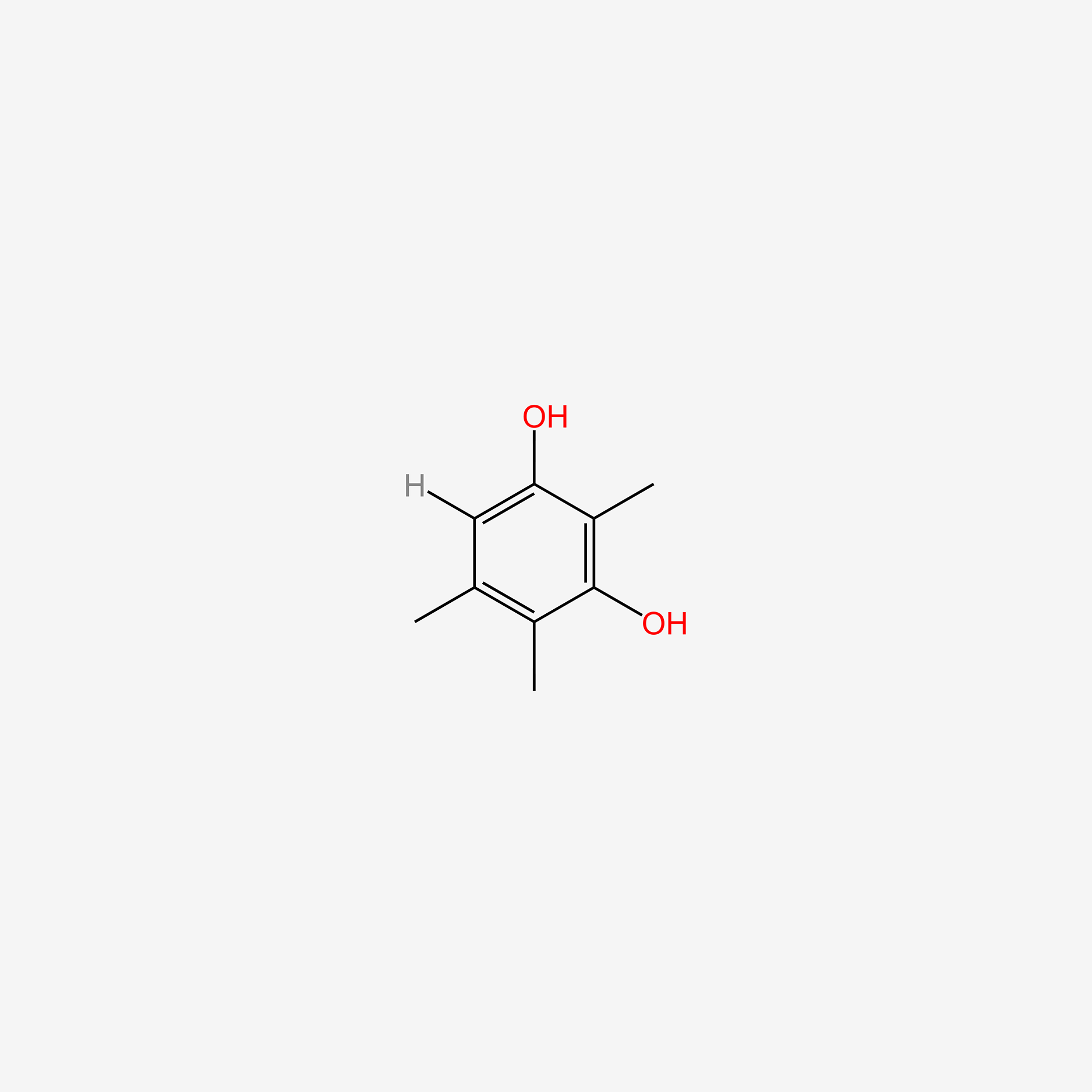 |
0.227 | D0C6DT | 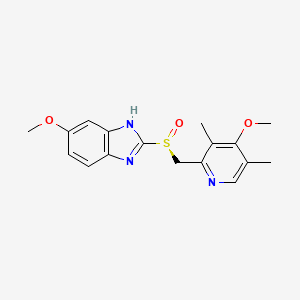 |
0.163 | ||
| ENC004240 |  |
0.222 | D01XNB |  |
0.163 | ||