NPs Basic Information
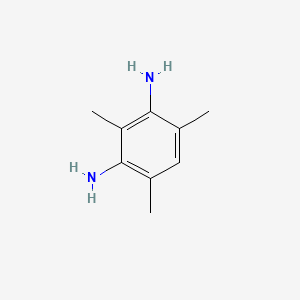
|
Name |
2,4-Diaminomesitylene
|
| Molecular Formula | C9H14N2 | |
| IUPAC Name* |
2,4,6-trimethylbenzene-1,3-diamine
|
|
| SMILES |
CC1=CC(=C(C(=C1N)C)N)C
|
|
| InChI |
InChI=1S/C9H14N2/c1-5-4-6(2)9(11)7(3)8(5)10/h4H,10-11H2,1-3H3
|
|
| InChIKey |
ZVDSMYGTJDFNHN-UHFFFAOYSA-N
|
|
| Synonyms |
3102-70-3; 2,4-Diaminomesitylene; 2,4,6-Trimethylbenzene-1,3-diamine; 2,4,6-Trimethyl-1,3-phenylenediamine; 2,4,6-Trimethyl-m-phenylenediamine; 1,3-Benzenediamine, 2,4,6-trimethyl-; 2,4-Mesitylenediamine; 2,4,6-Trimethyl-1,3-benzenediamine; 9K3JRF5932; NSC-10392; 1,3-diamino-2,4,6-trimethylbenzene; 2,4-Diamino-1,3,5-trimethylbenzene; CCRIS 6520; EINECS 221-456-9; BRN 2717221; NSC10392; 3MPDA; Cambridge id 5180400; 1, 2,4,6-trimethyl-; 3-13-00-00343 (Beilstein Handbook Reference); SCHEMBL262306; UNII-9K3JRF5932; CHEMBL1998053; 2,6-Trimethyl-m-phenylenediamine; DTXSID40184994; ZINC391994; m-Phenylenediamine,4,6-trimethyl-; ACT07451; ALBB-014271; MFCD00010149; NSC 10392; m-Phenylenediamine,2,4,6-trimethyl-; AKOS005174343; 2,4,6-Trimethyl-1,3-benzenediamine #; AS-12571; NCI60_000097; 2,4,6-Trimethyl-m-phenylenediamine, 96%; DB-005852; CS-0145866; FT-0609973; T1275; 1,3,5-TRIMETHYL-2,6-BENZENEDIAMINE; D83806; EN300-7366611; A820695; Q27894392; 858865-47-1
|
|
| CAS | 3102-70-3 | |
| PubChem CID | 76547 | |
| ChEMBL ID | CHEMBL1998053 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 150.22 | ALogp: | 1.7 |
| HBD: | 2 | HBA: | 2 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 52.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 11 | QED Weighted: | 0.558 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.637 | MDCK Permeability: | 0.00013065 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.685 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.075 |
| 30% Bioavailability (F30%): | 0.693 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.577 | Plasma Protein Binding (PPB): | 52.29% |
| Volume Distribution (VD): | 1.369 | Fu: | 62.21% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.272 | CYP1A2-substrate: | 0.754 |
| CYP2C19-inhibitor: | 0.055 | CYP2C19-substrate: | 0.909 |
| CYP2C9-inhibitor: | 0.013 | CYP2C9-substrate: | 0.124 |
| CYP2D6-inhibitor: | 0.161 | CYP2D6-substrate: | 0.83 |
| CYP3A4-inhibitor: | 0.017 | CYP3A4-substrate: | 0.307 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.062 | Half-life (T1/2): | 0.322 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.007 | Human Hepatotoxicity (H-HT): | 0.583 |
| Drug-inuced Liver Injury (DILI): | 0.37 | AMES Toxicity: | 0.166 |
| Rat Oral Acute Toxicity: | 0.058 | Maximum Recommended Daily Dose: | 0.929 |
| Skin Sensitization: | 0.869 | Carcinogencity: | 0.441 |
| Eye Corrosion: | 0.263 | Eye Irritation: | 0.99 |
| Respiratory Toxicity: | 0.49 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000181 | 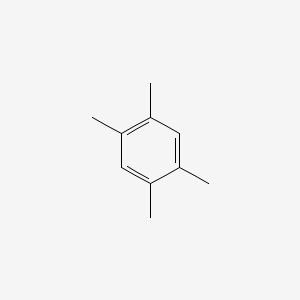 |
0.385 | D0FA2O | 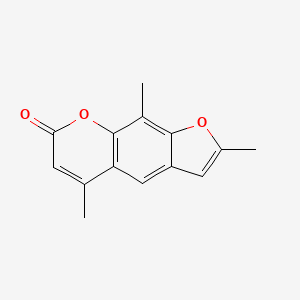 |
0.250 | ||
| ENC002336 | 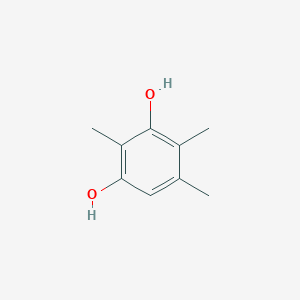 |
0.366 | D0X0RI | 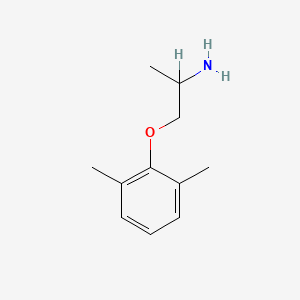 |
0.235 | ||
| ENC005230 | 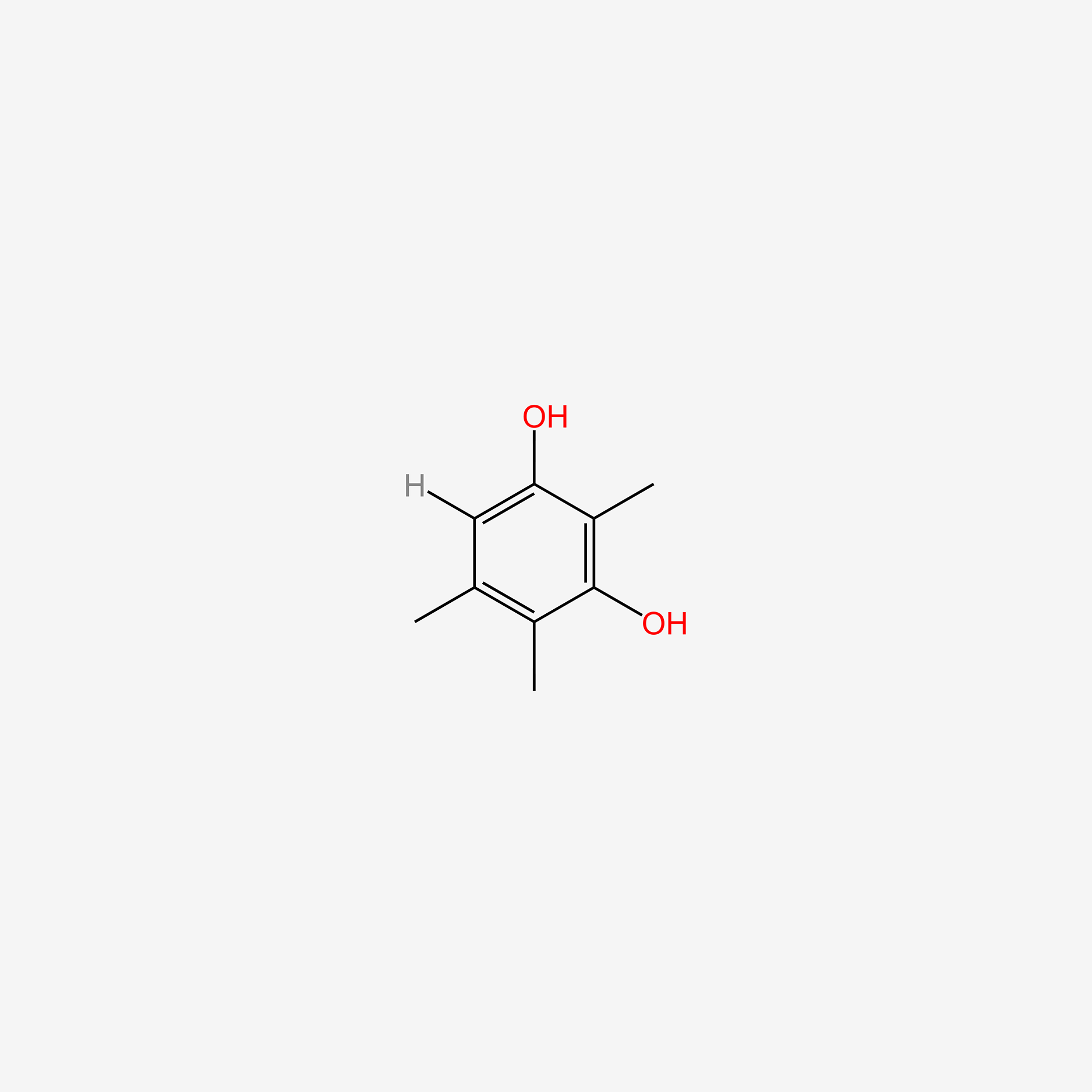 |
0.366 | D01PJR | 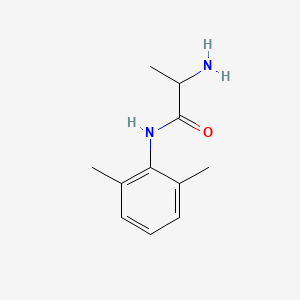 |
0.226 | ||
| ENC000342 | 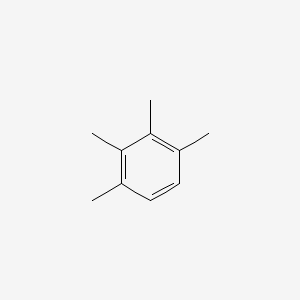 |
0.350 | D0Y4DY |  |
0.222 | ||
| ENC000180 |  |
0.333 | D09EBS |  |
0.215 | ||
| ENC000364 |  |
0.333 | D0L5FY |  |
0.211 | ||
| ENC001498 | 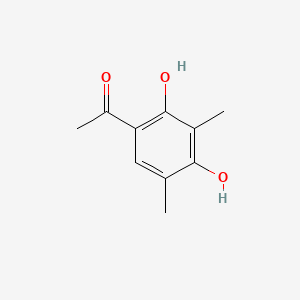 |
0.298 | D0U3DU | 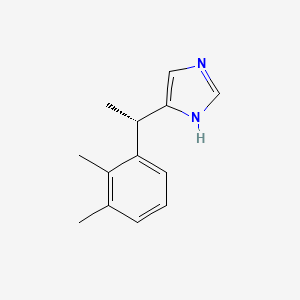 |
0.207 | ||
| ENC001026 | 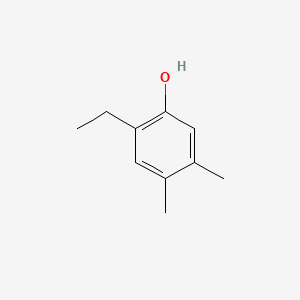 |
0.295 | D05YBZ | 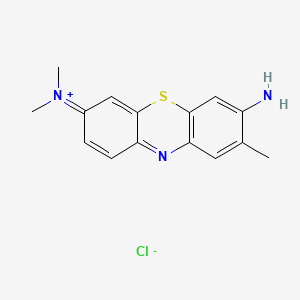 |
0.206 | ||
| ENC000084 | 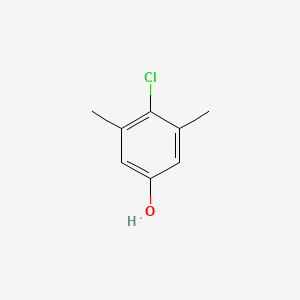 |
0.286 | D09TBD |  |
0.203 | ||
| ENC001359 | 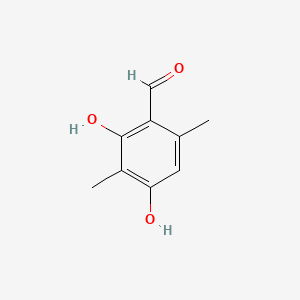 |
0.283 | D0X5NX | 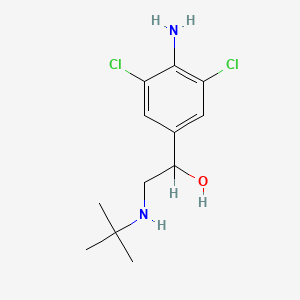 |
0.200 | ||