NPs Basic Information

|
Name |
Retene
|
| Molecular Formula | C18H18 | |
| IUPAC Name* |
1-methyl-7-propan-2-ylphenanthrene
|
|
| SMILES |
CC1=C2C=CC3=C(C2=CC=C1)C=CC(=C3)C(C)C
|
|
| InChI |
InChI=1S/C18H18/c1-12(2)14-7-10-17-15(11-14)8-9-16-13(3)5-4-6-18(16)17/h4-12H,1-3H3
|
|
| InChIKey |
NXLOLUFNDSBYTP-UHFFFAOYSA-N
|
|
| Synonyms |
RETENE; 483-65-8; 7-Isopropyl-1-methylphenanthrene; Reten; 1-Methyl-7-isopropylphenanthrene; Phenanthrene, 1-methyl-7-(1-methylethyl)-; 1-Methyl-7-(1-methylethyl)phenanthrene; Phenanthrene, 7-isopropyl-1-methyl-; 1-methyl-7-propan-2-ylphenanthrene; NSC 26317; 0W2D2E1P9Q; NCI-C55390; NSC-26317; CCRIS 3180; EINECS 207-597-9; UNII-0W2D2E1P9Q; AI3-00840; 1-methyl-7-(propan-2-yl)phenanthrene; RETENE [MI]; 1-methyl-7-isopropylphenathrene; DTXSID7058701; WLN: L B666J EY K1; Methyl-1-isopropyl-7-phenanthrene; NSC26317; ZINC1628335; Retene 10 microg/mL in Cyclohexane; MFCD00016358; DB-051547; FT-0632195; Q7316667
|
|
| CAS | 483-65-8 | |
| PubChem CID | 10222 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 234.3 | ALogp: | 6.5 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 3 |
| Heavy Atoms: | 18 | QED Weighted: | 0.477 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.711 | MDCK Permeability: | 0.00001320 |
| Pgp-inhibitor: | 0.431 | Pgp-substrate: | 0.078 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.888 |
| 30% Bioavailability (F30%): | 0.995 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.415 | Plasma Protein Binding (PPB): | 98.29% |
| Volume Distribution (VD): | 1.68 | Fu: | 0.93% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.919 | CYP1A2-substrate: | 0.641 |
| CYP2C19-inhibitor: | 0.611 | CYP2C19-substrate: | 0.334 |
| CYP2C9-inhibitor: | 0.468 | CYP2C9-substrate: | 0.873 |
| CYP2D6-inhibitor: | 0.774 | CYP2D6-substrate: | 0.927 |
| CYP3A4-inhibitor: | 0.391 | CYP3A4-substrate: | 0.375 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.519 | Half-life (T1/2): | 0.056 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.082 | Human Hepatotoxicity (H-HT): | 0.047 |
| Drug-inuced Liver Injury (DILI): | 0.901 | AMES Toxicity: | 0.867 |
| Rat Oral Acute Toxicity: | 0.193 | Maximum Recommended Daily Dose: | 0.873 |
| Skin Sensitization: | 0.893 | Carcinogencity: | 0.844 |
| Eye Corrosion: | 0.097 | Eye Irritation: | 0.995 |
| Respiratory Toxicity: | 0.086 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000098 |  |
0.438 | D0J6WW | 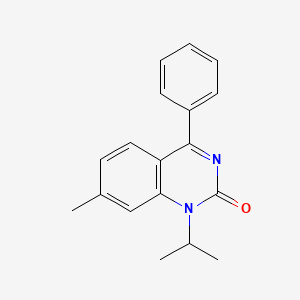 |
0.358 | ||
| ENC000167 | 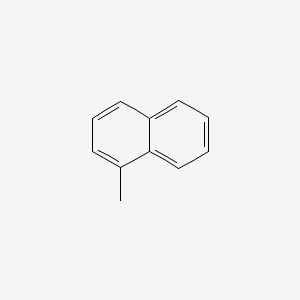 |
0.407 | D0DJ1B |  |
0.324 | ||
| ENC000368 | 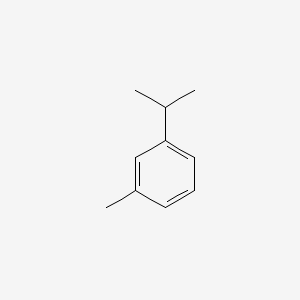 |
0.393 | D0R2OA | 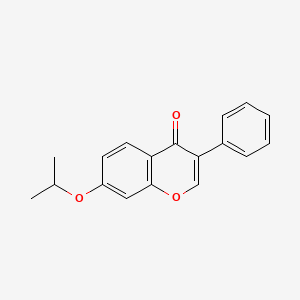 |
0.321 | ||
| ENC000347 |  |
0.379 | D0EL2O |  |
0.321 | ||
| ENC000199 |  |
0.368 | D0IT2X |  |
0.316 | ||
| ENC000365 |  |
0.322 | D02NTO |  |
0.310 | ||
| ENC000891 |  |
0.318 | D0H5LK |  |
0.297 | ||
| ENC000169 |  |
0.317 | D04JEE |  |
0.296 | ||
| ENC000392 |  |
0.308 | D0A1PX |  |
0.295 | ||
| ENC001388 |  |
0.307 | D05FTJ |  |
0.295 | ||