NPs Basic Information
|
Name |
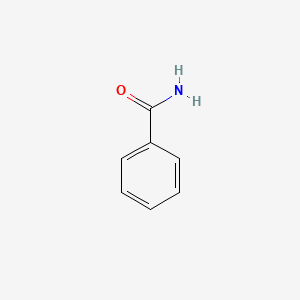
Benzamide
|
| Molecular Formula | C7H7NO | |
| IUPAC Name* |
benzamide
|
|
| SMILES |
C1=CC=C(C=C1)C(=O)N
|
|
| InChI |
InChI=1S/C7H7NO/c8-7(9)6-4-2-1-3-5-6/h1-5H,(H2,8,9)
|
|
| InChIKey |
KXDAEFPNCMNJSK-UHFFFAOYSA-N
|
|
| Synonyms |
Benzamide; 55-21-0; Benzoylamide; Benzoic acid amide; Phenylcarboxyamide; Phenylcarboxamide; Benzenecarboxamide; Amid kyseliny benzoove; Amid kyseliny benzoove [Czech]; NSC 3114; Phenyl Carboxyamide; BENZOIC ACID,AMIDE; MFCD00007968; CHEMBL267373; CHEBI:28179; NSC-3114; 55738-52-8; 6X80438640; CCRIS 4594; HSDB 6360; EINECS 200-227-7; benzeneamide; benzimide; BRN 0385876; phenylamide; N-benzoylamine; benzoyl nitrogen; AI3-01031; benzene carboxamide; benzene-carboxamide; Benzamide, 99%; UNII-6X80438640; BENZAMIDE [MI]; BENZAMIDE [HSDB]; WLN: ZVR; benzene carboximidoic acid; PhC(O)NH2; Lopac-B-2009; DSSTox_CID_1709; bmse000668; PhC(=O)NH2; DSSTox_RID_76288; DSSTox_GSID_21709; Lopac0_000160; SCHEMBL16352; 4-09-00-00725 (Beilstein Handbook Reference); benzamide (ACD/Name 4.0); MLS000069472; Benzamide, p.a., 98.0%; DTXSID0021709; NSC3114; HMS2231M11; HMS3260O22; HMS3371I16; HMS3885L18; ZINC152996; CS-Z0019; HY-Z0283; Tox21_200621; Tox21_500160; BDBM50106187; s4715; STK069333; AKOS000118773; CCG-204255; LP00160; SDCCGSBI-0050148.P002; CAS-55-21-0; Benzamide, purum, >=98.0% (HPLC); NCGC00015142-01; NCGC00015142-02; NCGC00015142-03; NCGC00015142-04; NCGC00015142-05; NCGC00015142-06; NCGC00015142-07; NCGC00091355-01; NCGC00091355-02; NCGC00091355-03; NCGC00258175-01; NCGC00260845-01; BP-21224; DS-17194; SMR000059089; SY047098; Benzamide, Vetec(TM) reagent grade, 98%; B0012; B0220; B1418; EU-0100160; FT-0622630; FT-0622631; EN300-15618; B 2009; Benzamide, purified by sublimation, >=99.5%; C09815; D70176; A830526; Q417731; SR-01000075601; SR-01000075601-1; Z33546506; Benzamide, zone-refined, purified by sublimation, 99.9%; F3145-2903; Sulfabenzamide|Sulfabenzid|Sulfabenzide|Sulfabenzoylamide|N-Sulfamylbenzamide
|
|
| CAS | 55-21-0 | |
| PubChem CID | 2331 | |
| ChEMBL ID | CHEMBL267373 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 121.14 | ALogp: | 0.6 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 43.1 | Aromatic Rings: | 1 |
| Heavy Atoms: | 9 | QED Weighted: | 0.596 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.596 | MDCK Permeability: | 0.00003810 |
| Pgp-inhibitor: | 0.038 | Pgp-substrate: | 0.947 |
| Human Intestinal Absorption (HIA): | 0.008 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.773 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.998 | Plasma Protein Binding (PPB): | 67.70% |
| Volume Distribution (VD): | 1.182 | Fu: | 47.38% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.668 | CYP1A2-substrate: | 0.288 |
| CYP2C19-inhibitor: | 0.074 | CYP2C19-substrate: | 0.064 |
| CYP2C9-inhibitor: | 0.023 | CYP2C9-substrate: | 0.133 |
| CYP2D6-inhibitor: | 0.007 | CYP2D6-substrate: | 0.28 |
| CYP3A4-inhibitor: | 0.008 | CYP3A4-substrate: | 0.169 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 8.588 | Half-life (T1/2): | 0.413 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.102 | Human Hepatotoxicity (H-HT): | 0.071 |
| Drug-inuced Liver Injury (DILI): | 0.736 | AMES Toxicity: | 0.01 |
| Rat Oral Acute Toxicity: | 0.04 | Maximum Recommended Daily Dose: | 0.01 |
| Skin Sensitization: | 0.175 | Carcinogencity: | 0.113 |
| Eye Corrosion: | 0.016 | Eye Irritation: | 0.988 |
| Respiratory Toxicity: | 0.061 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000192 |  |
0.667 | D0X9RY |  |
0.667 | ||
| ENC000013 |  |
0.667 | D07ONP | 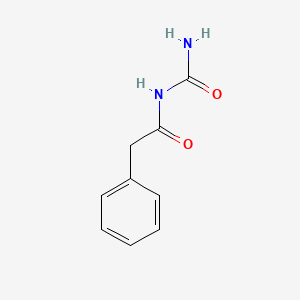 |
0.452 | ||
| ENC000651 |  |
0.618 | D0R1CR | 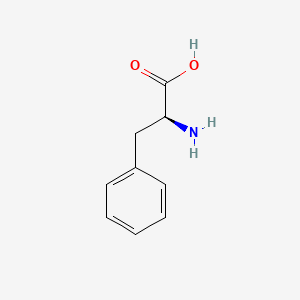 |
0.450 | ||
| ENC000174 | 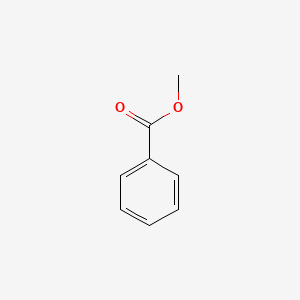 |
0.606 | D01ZJK | 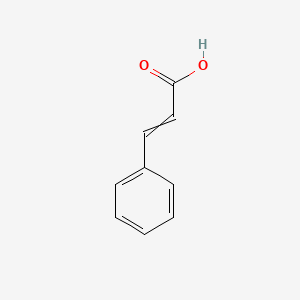 |
0.436 | ||
| ENC005854 |  |
0.559 | D0B7OD | 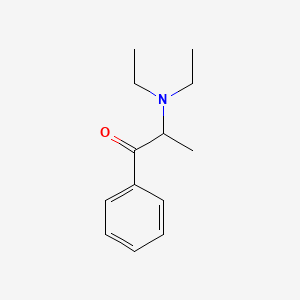 |
0.435 | ||
| ENC000219 | 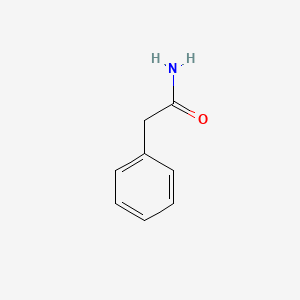 |
0.559 | D05OIS | 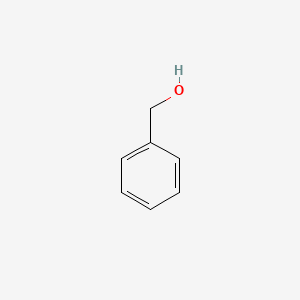 |
0.412 | ||
| ENC000175 | 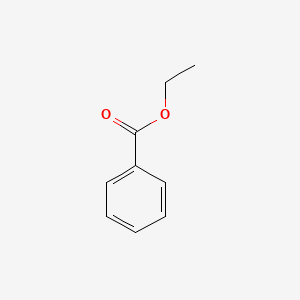 |
0.556 | D0P2GK | 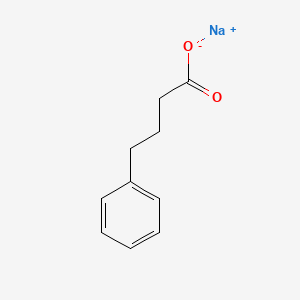 |
0.395 | ||
| ENC000637 | 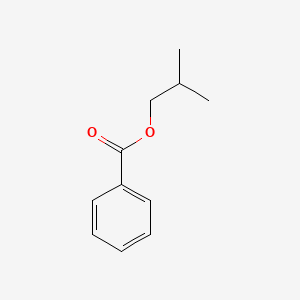 |
0.488 | D05BMG | 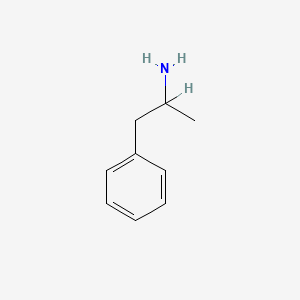 |
0.395 | ||
| ENC001049 | 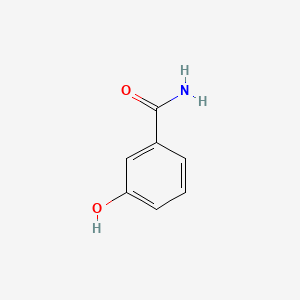 |
0.486 | D0H0HJ | 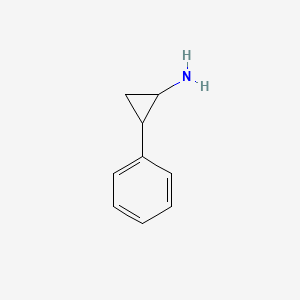 |
0.395 | ||
| ENC000108 | 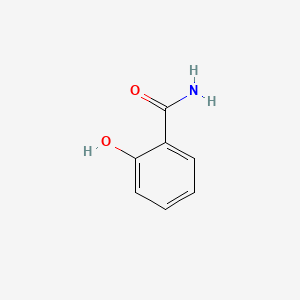 |
0.486 | D0T3LF | 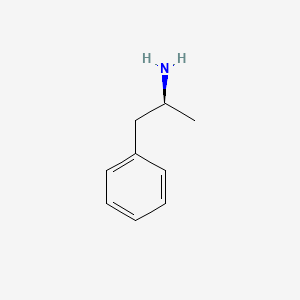 |
0.395 | ||