NPs Basic Information
|
Name |
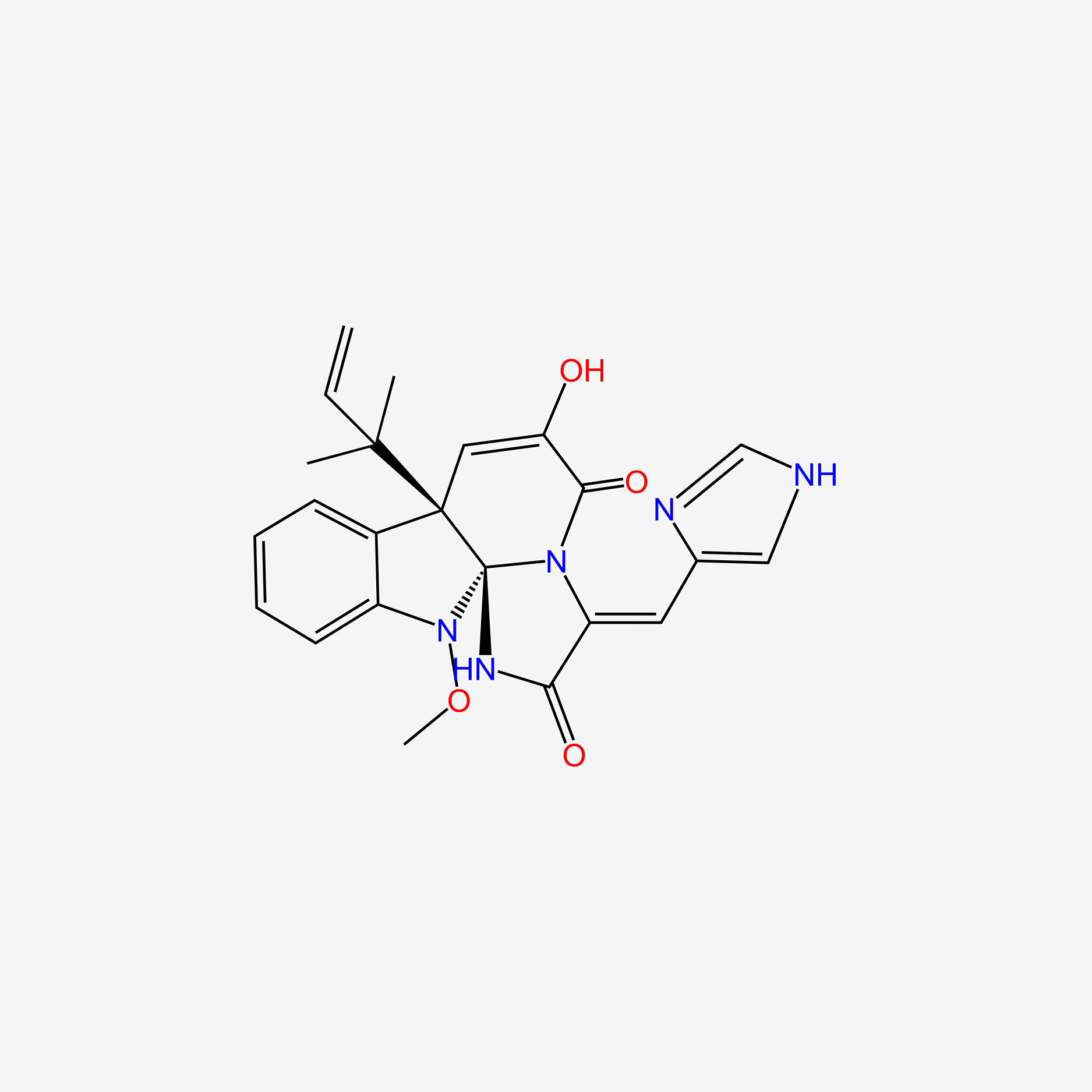
Isomeleagrin
|
| Molecular Formula | C23H23N5O4 | |
| IUPAC Name* |
11-hydroxy-14-(1H-imidazol-4-ylmethylidene)-2-methoxy-9-(2-methylbut-3-en-2-yl)-2,13,16-triazatetracyclo[7.7.0.01,13.03,8]hexadeca-3,5,7,10-tetraene-12,15-dione
|
|
| SMILES |
C=CC(C)(C)C12C=C(O)C(=O)N3C(=Cc4c[nH]cn4)C(=O)NC31N(OC)c1ccccc12
|
|
| InChI |
InChI=1S/C23H23N5O4/c1-5-21(2,3)22-11-18(29)20(31)27-17(10-14-12-24-13-25-14)19(30)26-23(22,27)28(32-4)16-9-7-6-8-15(16)22/h5-13,29H,1H2,2-4H3,(H,24,25)(H,26,30)/b17-10-/t22-,23-/m0/s1
|
|
| InChIKey |
JTJJJLSLKZFEPJ-WJFVXKAHSA-N
|
|
| Synonyms |
NA
|
|
| CAS | NA | |
| PubChem CID | NA | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was calculated by STOUT. Reference: PMID:33906675.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 433.47 | ALogp: | 2.3 |
| HBD: | 3 | HBA: | 6 |
| Rotatable Bonds: | 4 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 110.8 | Aromatic Rings: | 5 |
| Heavy Atoms: | 32 | QED Weighted: | 0.505 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.808 | MDCK Permeability: | 0.00002270 |
| Pgp-inhibitor: | 0.999 | Pgp-substrate: | 0.068 |
| Human Intestinal Absorption (HIA): | 0.909 | 20% Bioavailability (F20%): | 0.676 |
| 30% Bioavailability (F30%): | 0.983 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.231 | Plasma Protein Binding (PPB): | 67.67% |
| Volume Distribution (VD): | 0.394 | Fu: | 28.61% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.041 | CYP1A2-substrate: | 0.63 |
| CYP2C19-inhibitor: | 0.669 | CYP2C19-substrate: | 0.75 |
| CYP2C9-inhibitor: | 0.809 | CYP2C9-substrate: | 0.576 |
| CYP2D6-inhibitor: | 0.74 | CYP2D6-substrate: | 0.05 |
| CYP3A4-inhibitor: | 0.966 | CYP3A4-substrate: | 0.95 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.856 | Half-life (T1/2): | 0.231 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0 | Human Hepatotoxicity (H-HT): | 0.246 |
| Drug-inuced Liver Injury (DILI): | 0.972 | AMES Toxicity: | 0.867 |
| Rat Oral Acute Toxicity: | 0.954 | Maximum Recommended Daily Dose: | 0.18 |
| Skin Sensitization: | 0.814 | Carcinogencity: | 0.807 |
| Eye Corrosion: | 0.005 | Eye Irritation: | 0.119 |
| Respiratory Toxicity: | 0.819 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC002908 | 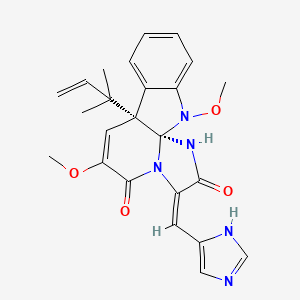 |
0.676 | D0W7WC | 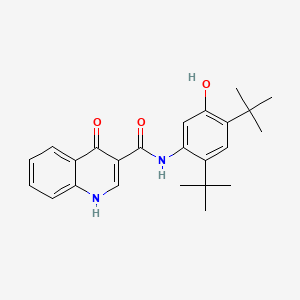 |
0.246 | ||
| ENC004493 | 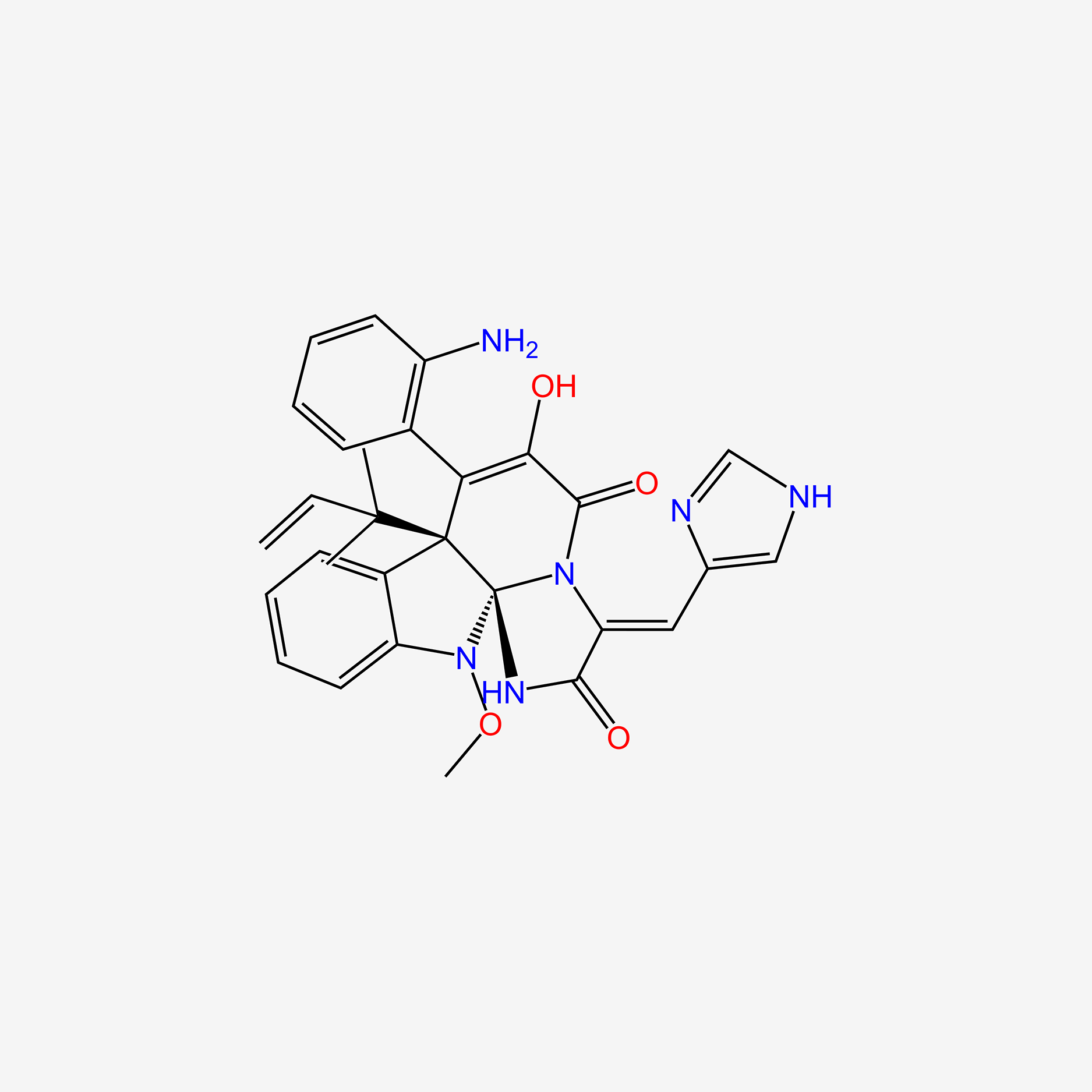 |
0.664 | D09NIA |  |
0.242 | ||
| ENC004494 | 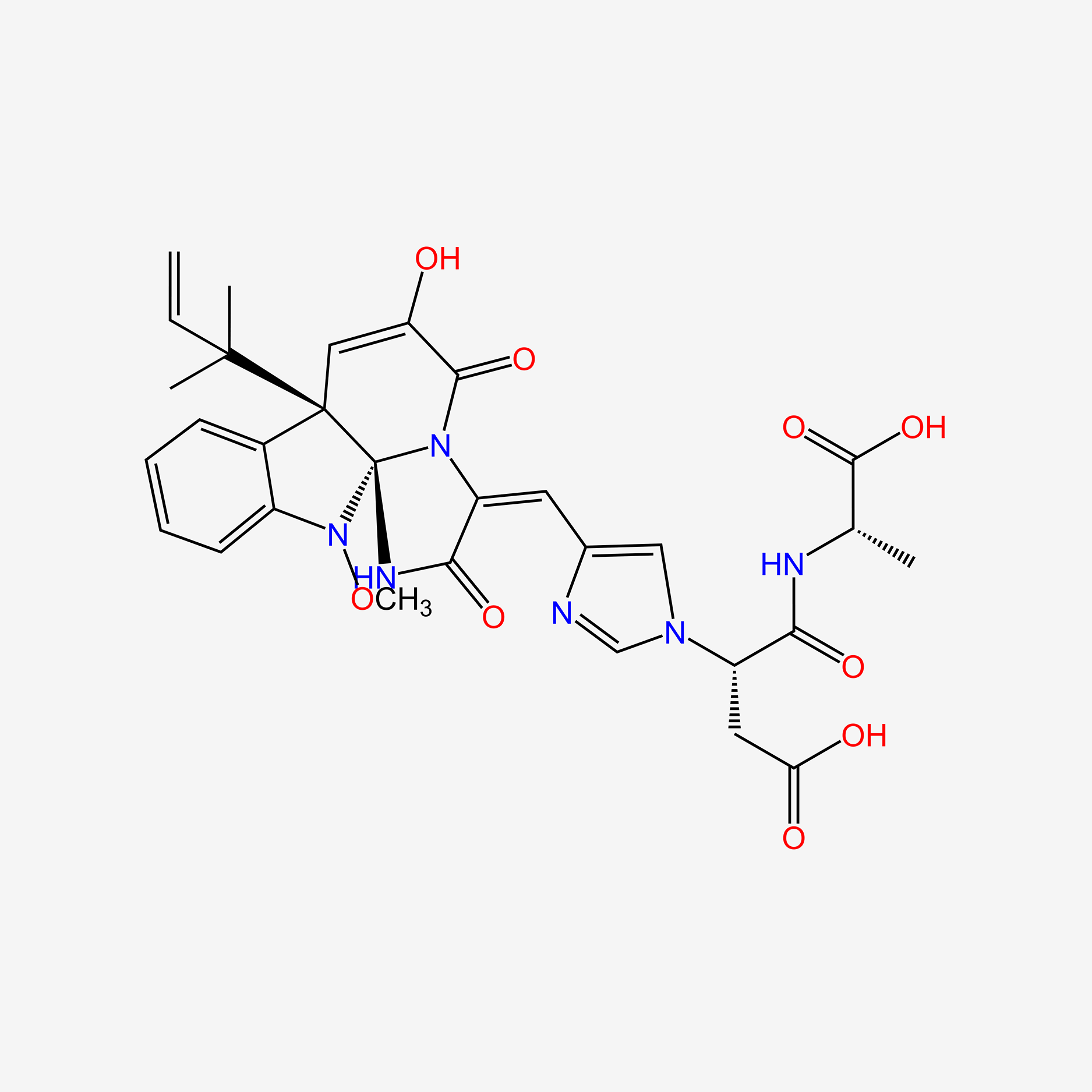 |
0.611 | D01PZD | 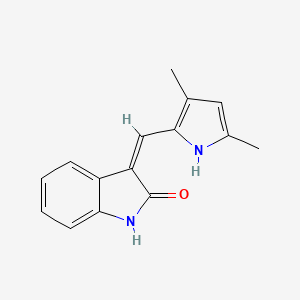 |
0.239 | ||
| ENC004495 | 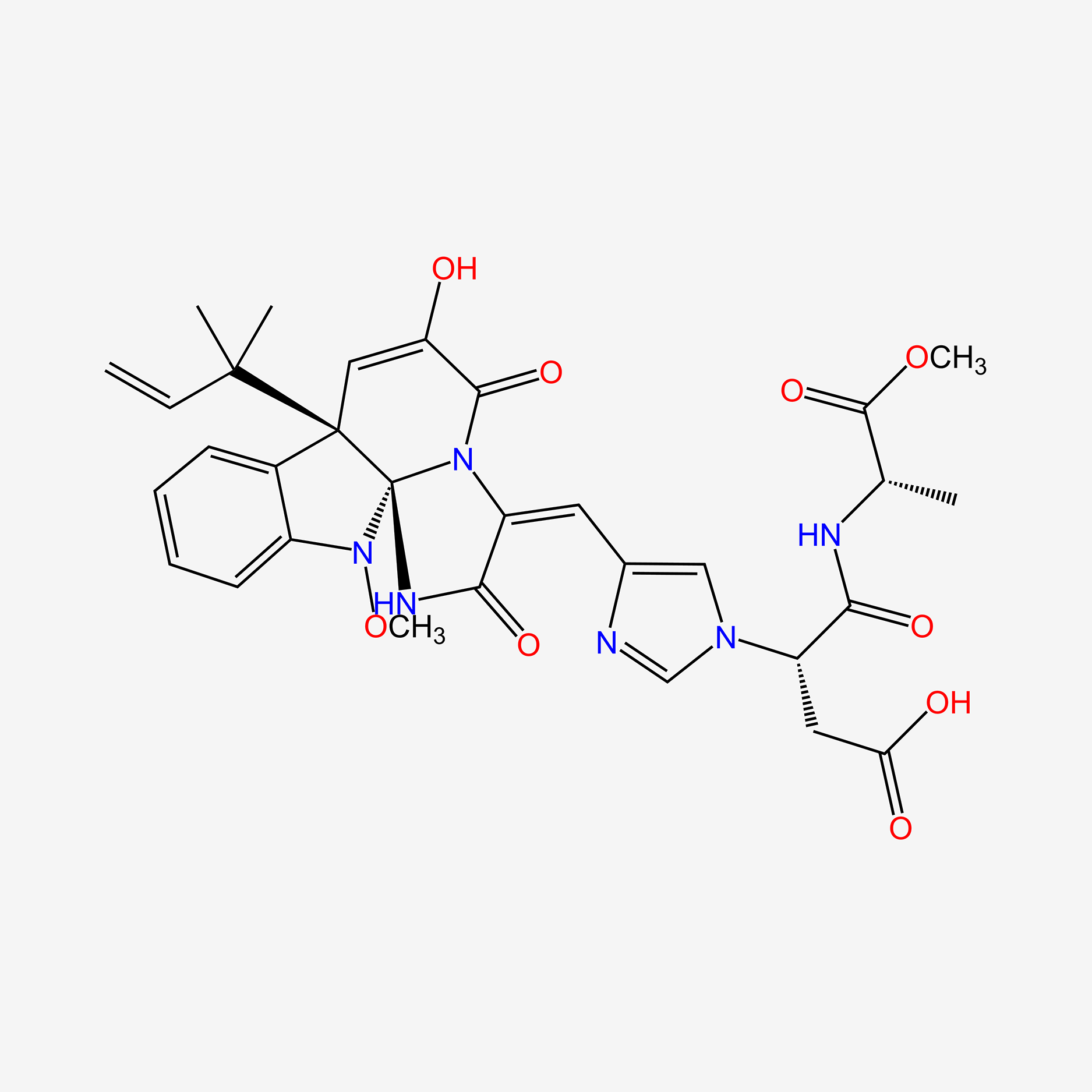 |
0.597 | D06GKN | 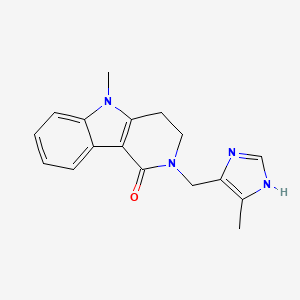 |
0.236 | ||
| ENC004496 | 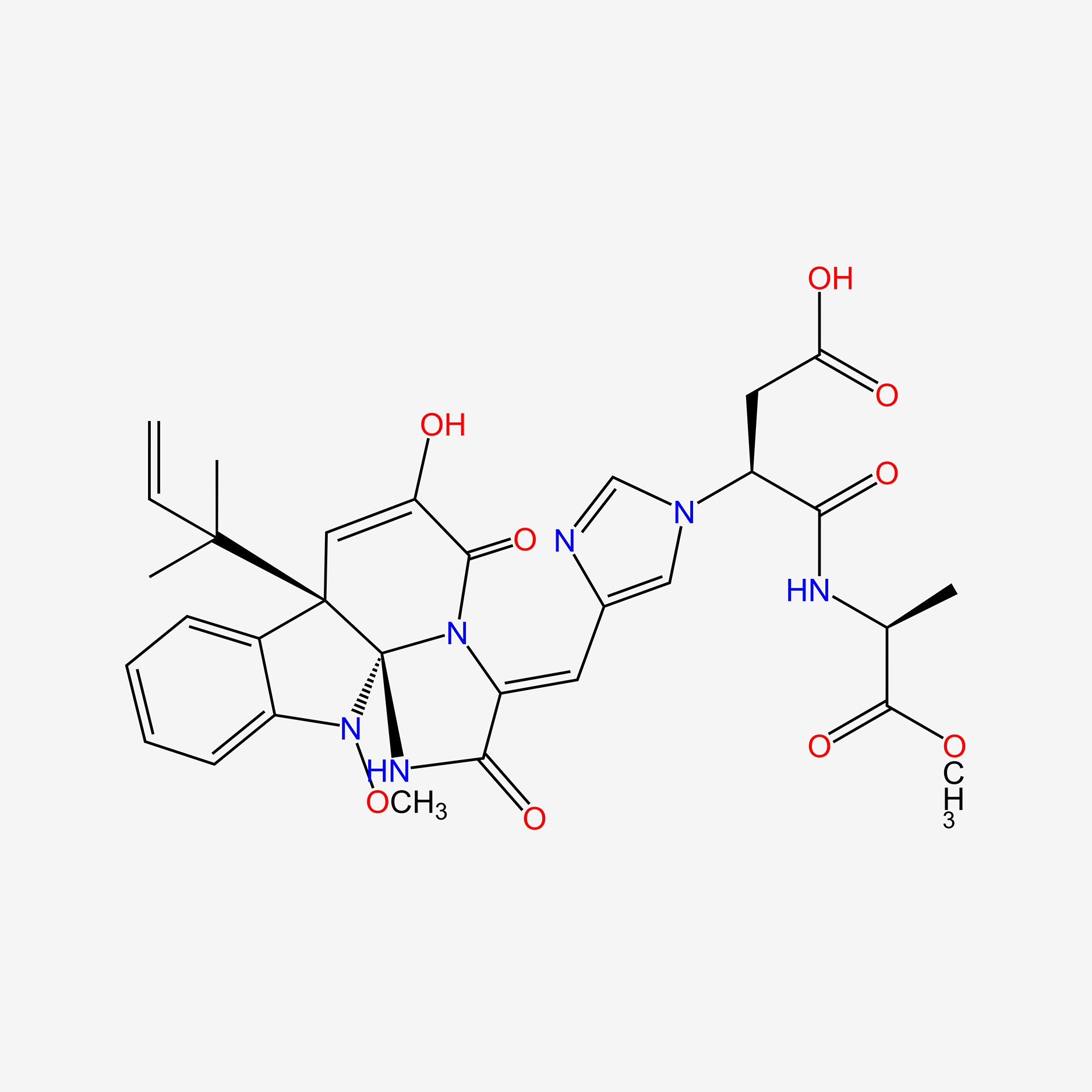 |
0.597 | D01WLC | 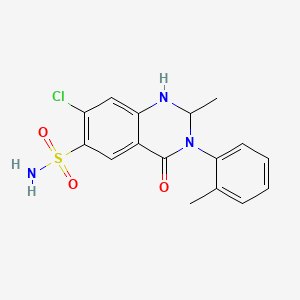 |
0.232 | ||
| ENC004497 | 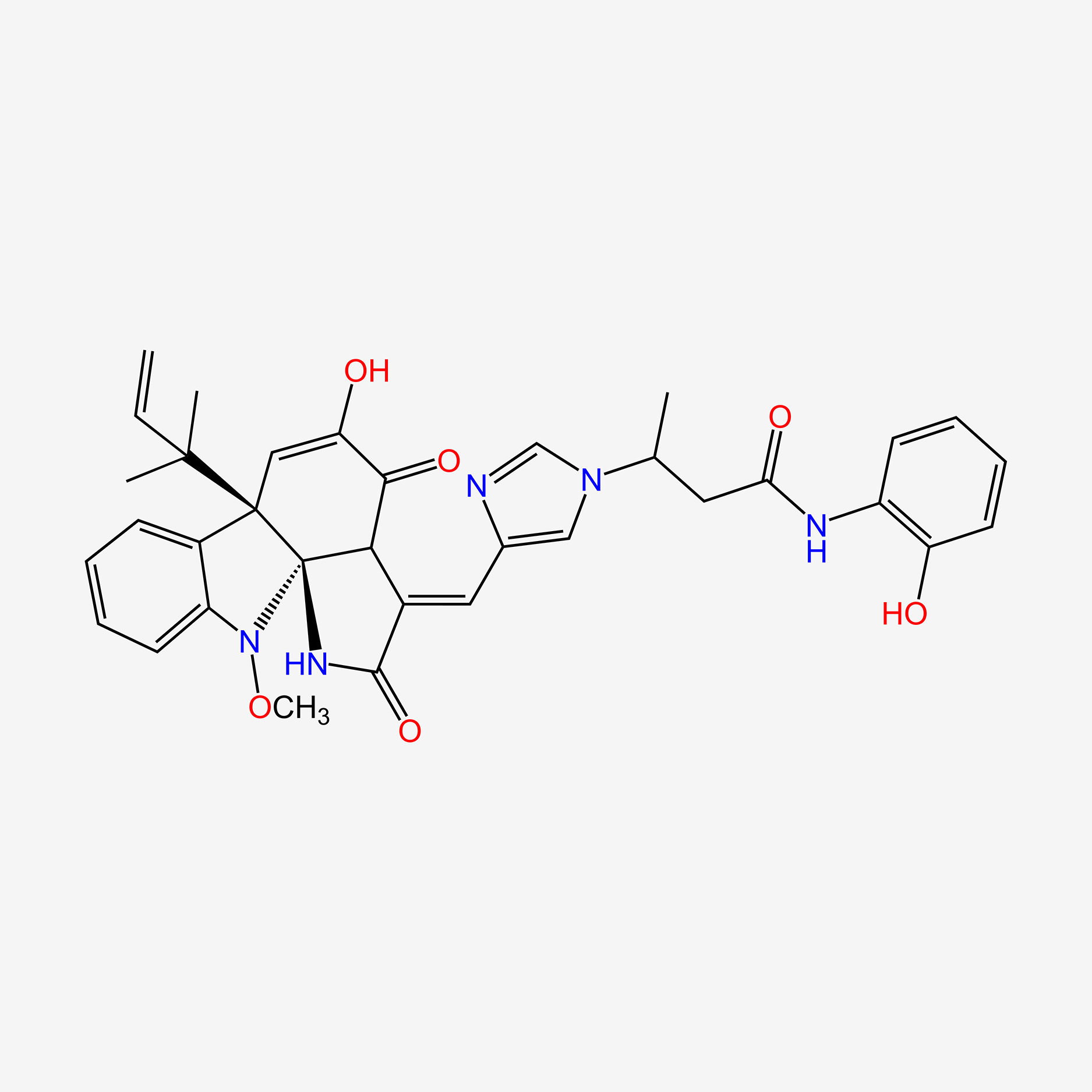 |
0.446 | D0T0LM | 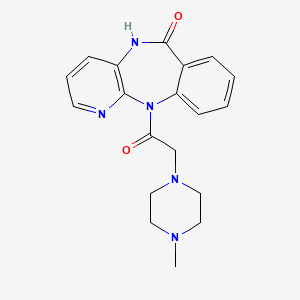 |
0.224 | ||
| ENC006112 |  |
0.365 | D09WKB |  |
0.222 | ||
| ENC003246 |  |
0.336 | D07RGW |  |
0.220 | ||
| ENC003221 |  |
0.336 | D0E3WQ |  |
0.216 | ||
| ENC002459 | 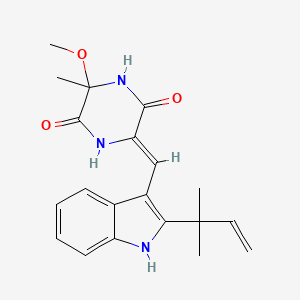 |
0.333 | D0W2NM | 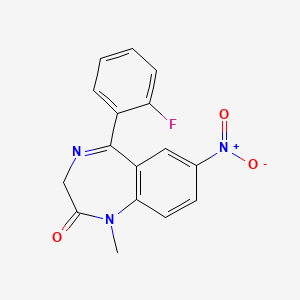 |
0.214 | ||