NPs Basic Information
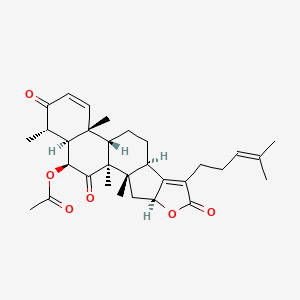
|
Name |
16-O-deacetylhelvolic acid 21,16-lactone
|
| Molecular Formula | C31H40O6 | |
| IUPAC Name* |
[(1S,2S,4S,9R,12S,13R,17S,18S,19S)-1,2,13,17-tetramethyl-7-(4-methylpent-3-enyl)-6,16,20-trioxo-5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icosa-7,14-dien-19-yl] acetate
|
|
| SMILES |
C[C@H]1[C@@H]2[C@@H](C(=O)[C@]3([C@H]([C@]2(C=CC1=O)C)CC[C@@H]4[C@@]3(C[C@H]5C4=C(C(=O)O5)CCC=C(C)C)C)C)OC(=O)C
|
|
| InChI |
InChI=1S/C31H40O6/c1-16(2)9-8-10-19-24-20-11-12-23-29(5)14-13-21(33)17(3)25(29)26(36-18(4)32)27(34)31(23,7)30(20,6)15-22(24)37-28(19)35/h9,13-14,17,20,22-23,25-26H,8,10-12,15H2,1-7H3/t17-,20+,22+,23+,25-,26+,29-,30+,31-/m1/s1
|
|
| InChIKey |
UJLGIFPRBONJKE-PREXDRGQSA-N
|
|
| Synonyms |
CHEMBL4211894; 16-O-deacetylhelvolic acid 21,16-lactone
|
|
| CAS | NA | |
| PubChem CID | 139590094 | |
| ChEMBL ID | CHEMBL4211894 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 508.6 | ALogp: | 5.3 |
| HBD: | 0 | HBA: | 6 |
| Rotatable Bonds: | 5 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 86.7 | Aromatic Rings: | 5 |
| Heavy Atoms: | 37 | QED Weighted: | 0.365 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.993 | MDCK Permeability: | 0.00002830 |
| Pgp-inhibitor: | 0.962 | Pgp-substrate: | 0.638 |
| Human Intestinal Absorption (HIA): | 0.028 | 20% Bioavailability (F20%): | 0.019 |
| 30% Bioavailability (F30%): | 0.059 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.963 | Plasma Protein Binding (PPB): | 98.75% |
| Volume Distribution (VD): | 2.119 | Fu: | 4.22% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.038 | CYP1A2-substrate: | 0.201 |
| CYP2C19-inhibitor: | 0.041 | CYP2C19-substrate: | 0.615 |
| CYP2C9-inhibitor: | 0.159 | CYP2C9-substrate: | 0.185 |
| CYP2D6-inhibitor: | 0.058 | CYP2D6-substrate: | 0.09 |
| CYP3A4-inhibitor: | 0.737 | CYP3A4-substrate: | 0.58 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.312 | Half-life (T1/2): | 0.069 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.012 | Human Hepatotoxicity (H-HT): | 0.753 |
| Drug-inuced Liver Injury (DILI): | 0.569 | AMES Toxicity: | 0.031 |
| Rat Oral Acute Toxicity: | 0.983 | Maximum Recommended Daily Dose: | 0.916 |
| Skin Sensitization: | 0.277 | Carcinogencity: | 0.853 |
| Eye Corrosion: | 0.004 | Eye Irritation: | 0.018 |
| Respiratory Toxicity: | 0.984 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC003847 | 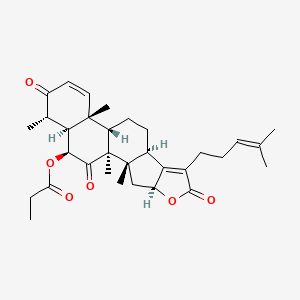 |
0.897 | D0X7XG | 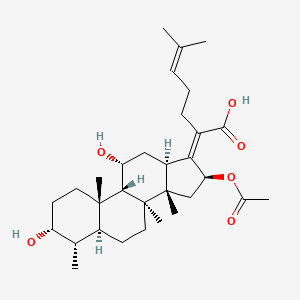 |
0.327 | ||
| ENC005152 | 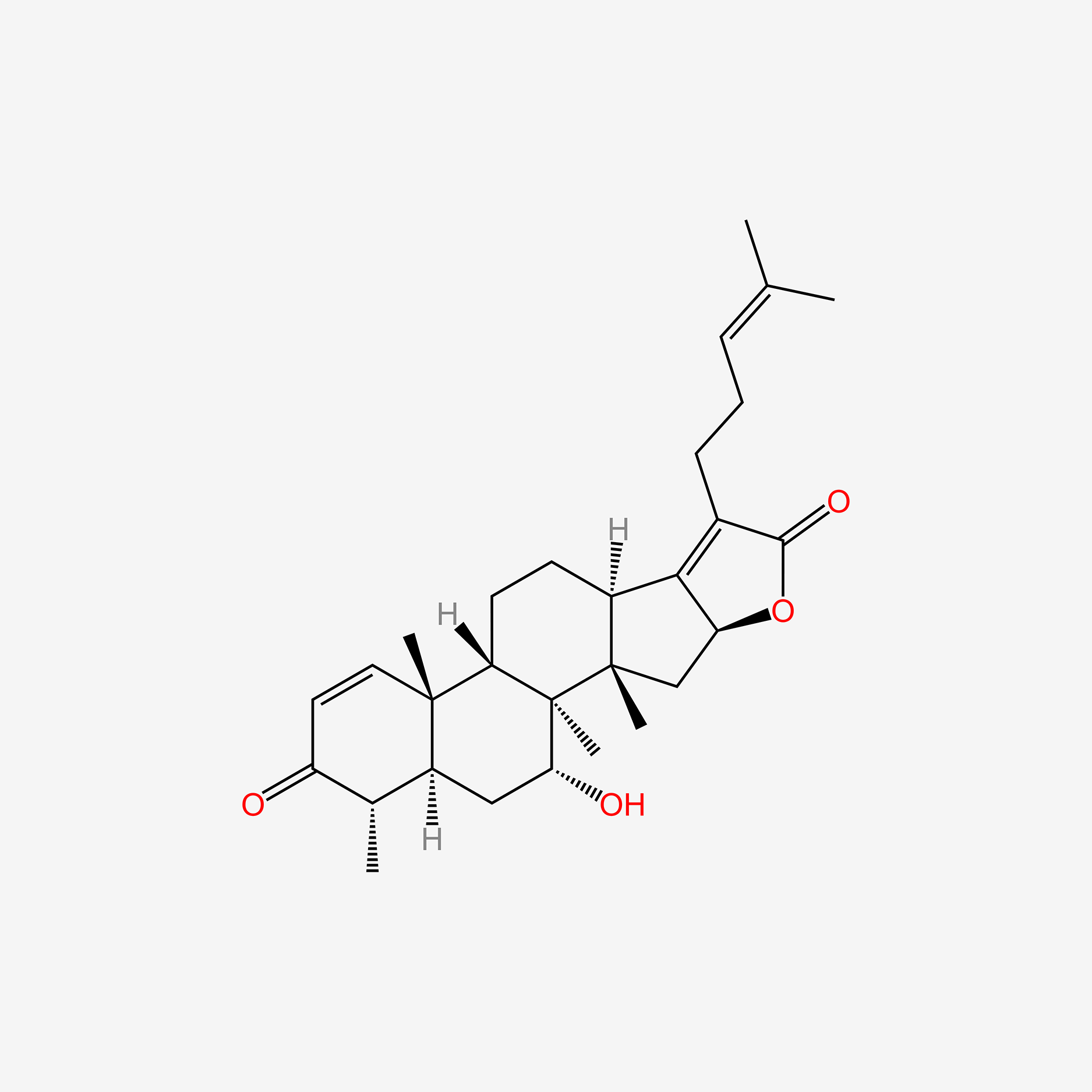 |
0.652 | D09WYX | 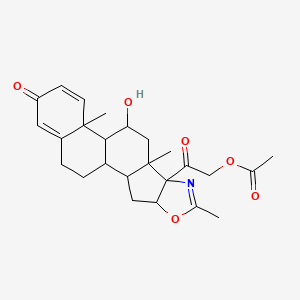 |
0.279 | ||
| ENC001480 | 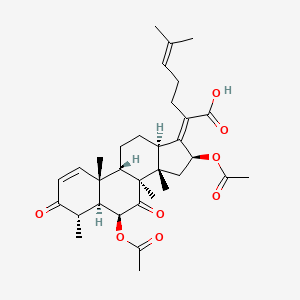 |
0.633 | D0D2TN | 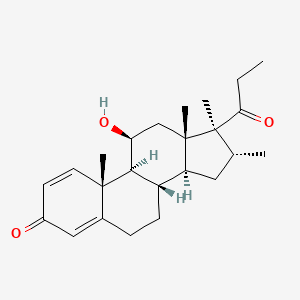 |
0.263 | ||
| ENC003484 | 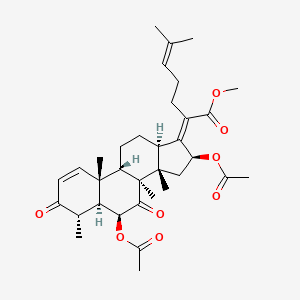 |
0.618 | D0G7KJ | 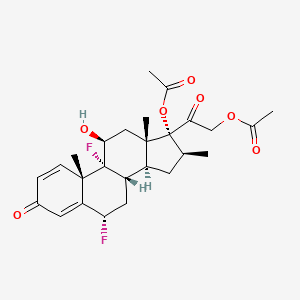 |
0.261 | ||
| ENC005487 | 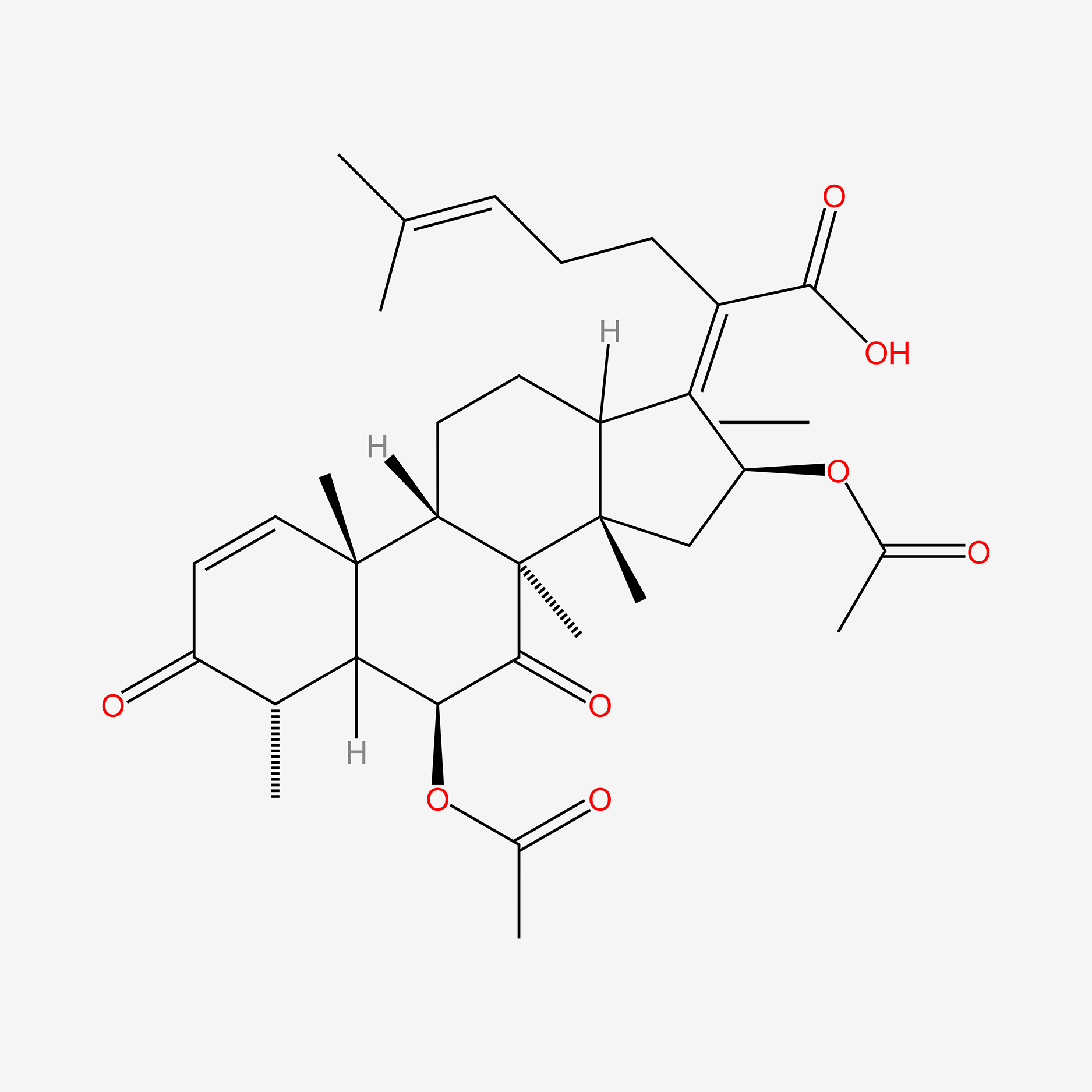 |
0.618 | D0E9KA | 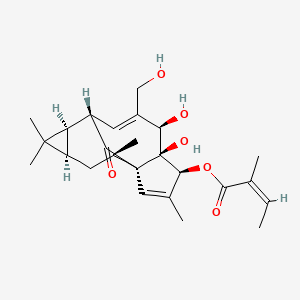 |
0.255 | ||
| ENC003780 | 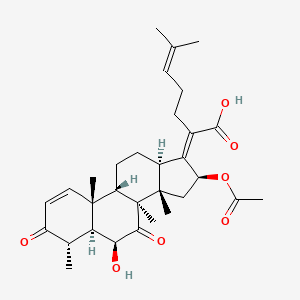 |
0.608 | D03ZZK | 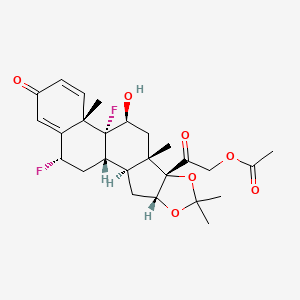 |
0.252 | ||
| ENC005151 | 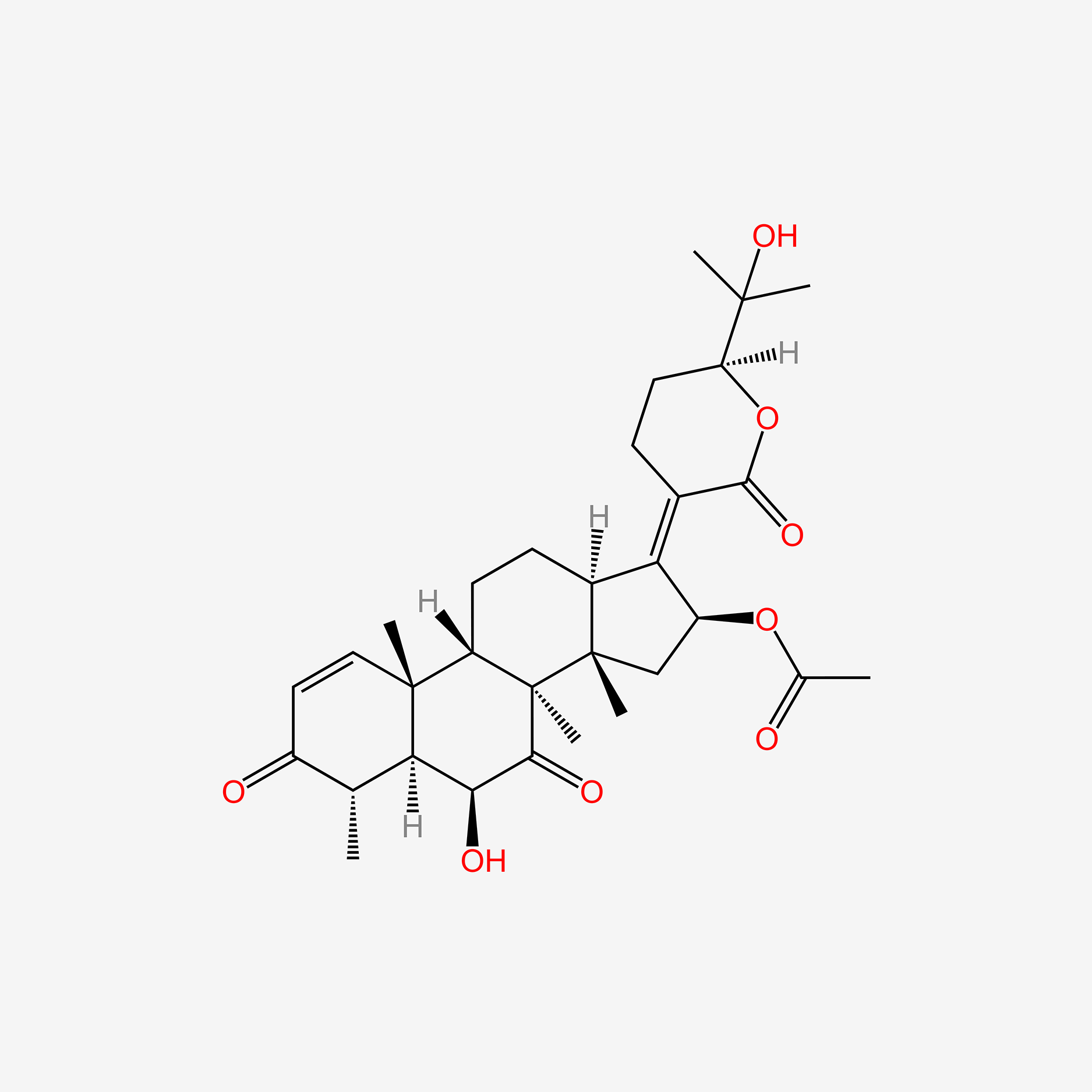 |
0.557 | D0W2EK | 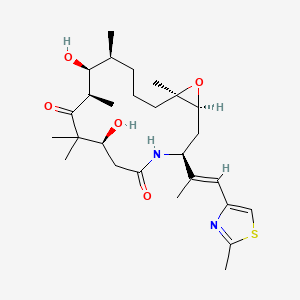 |
0.244 | ||
| ENC005155 | 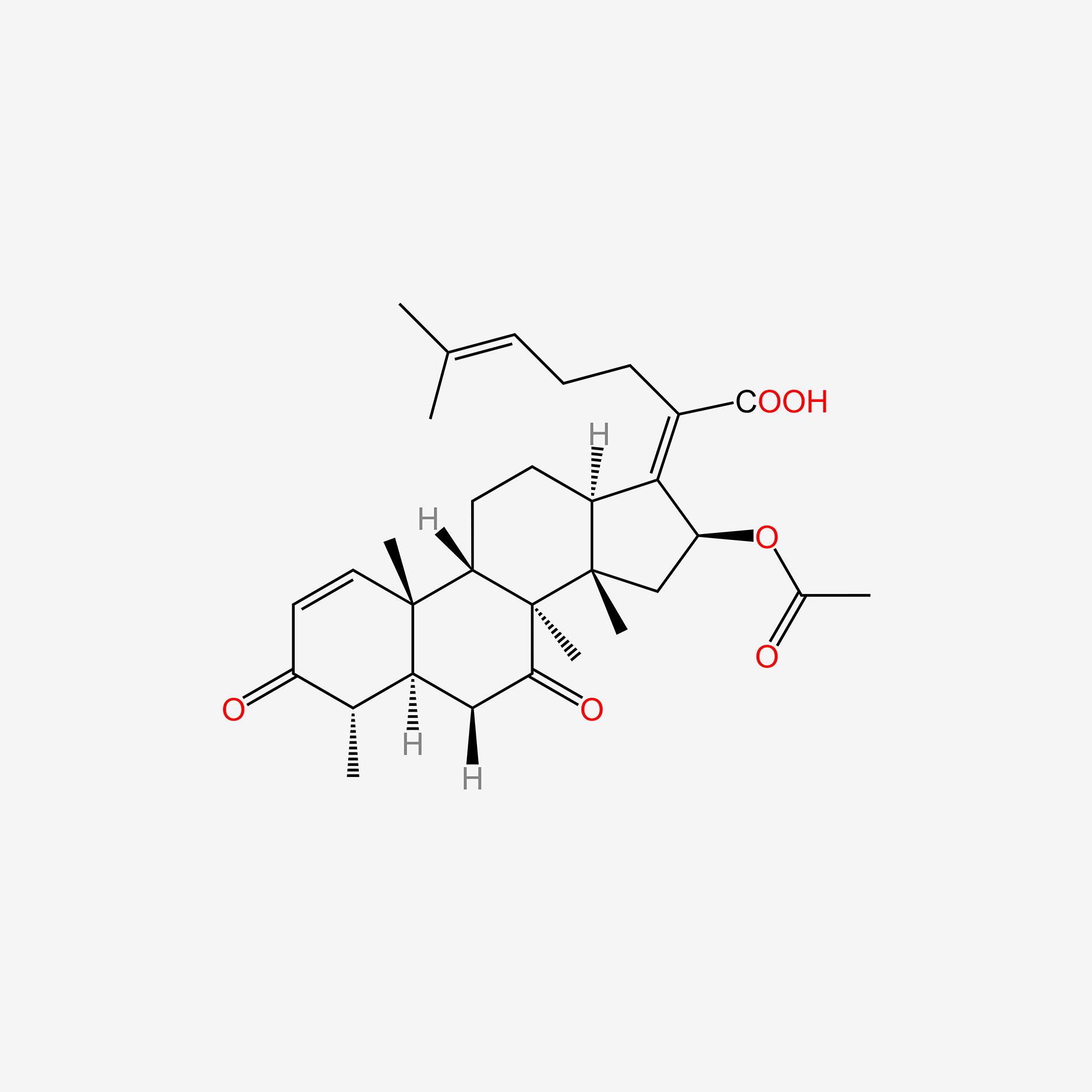 |
0.543 | D0H2MO | 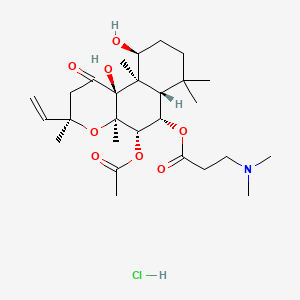 |
0.242 | ||
| ENC005236 | 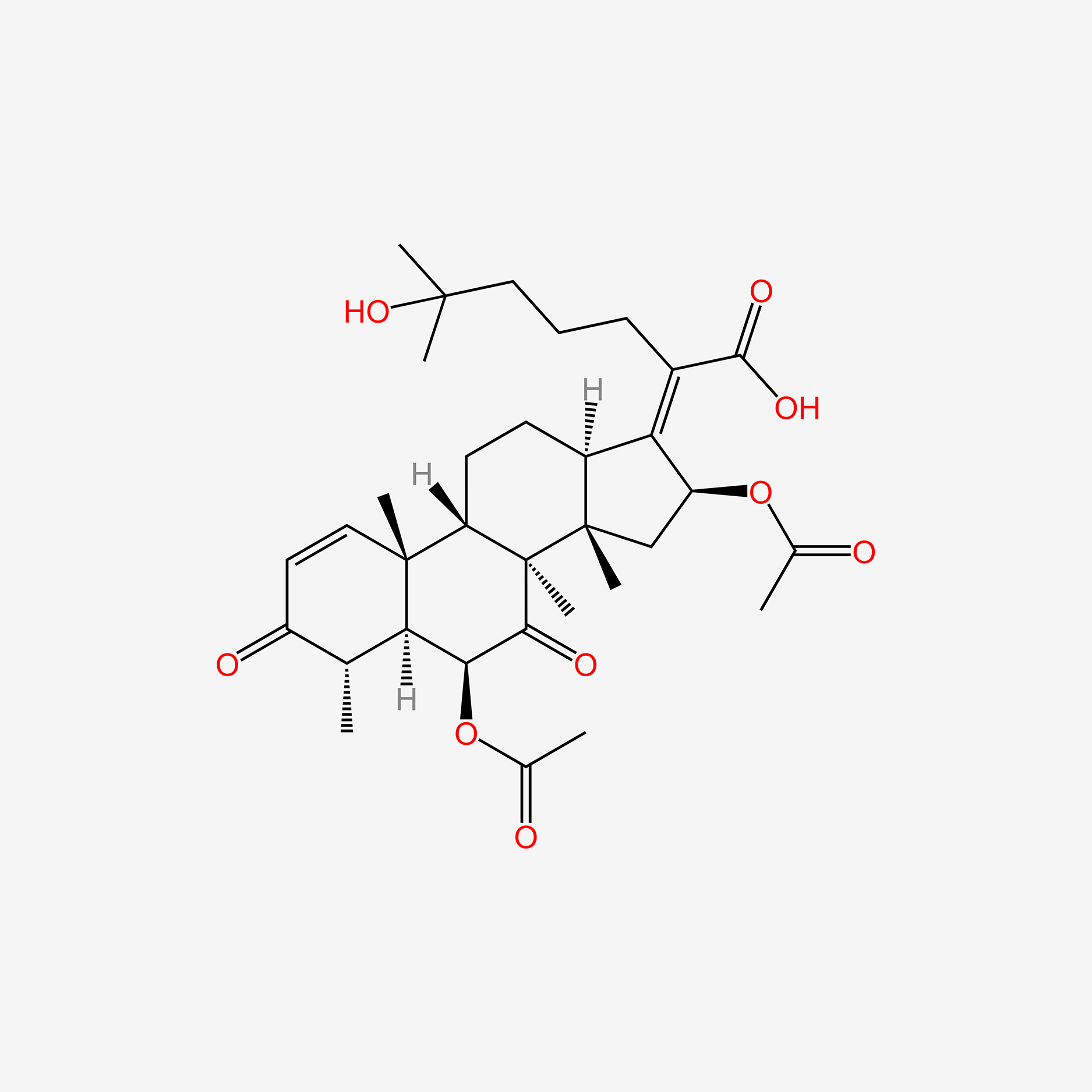 |
0.540 | D09IEE | 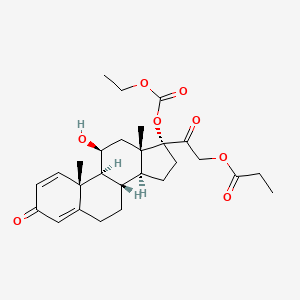 |
0.241 | ||
| ENC005154 | 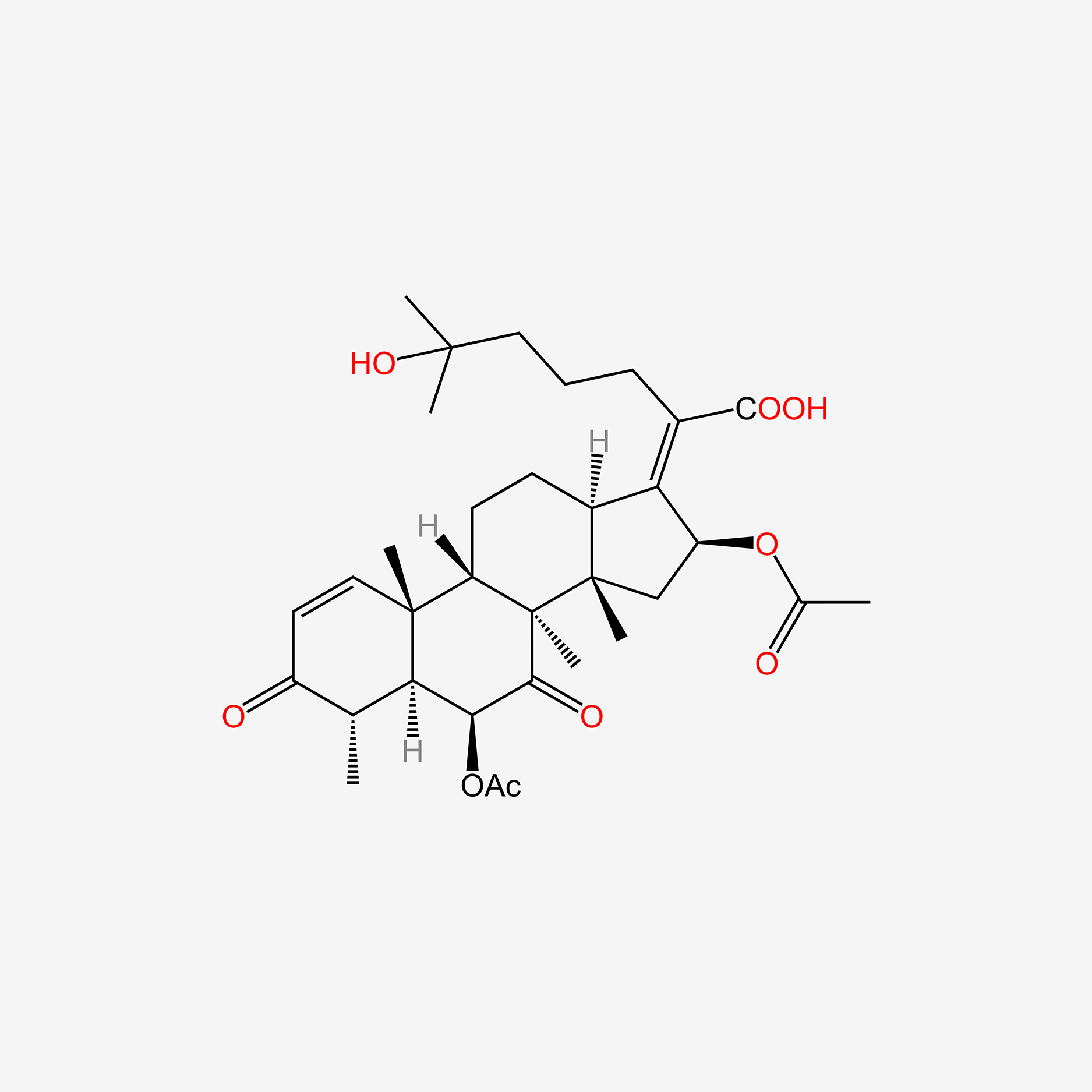 |
0.540 | D01ZOG |  |
0.239 | ||