NPs Basic Information
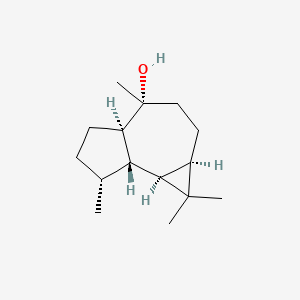
|
Name |
(-)-Globulol
|
| Molecular Formula | C15H26O | |
| IUPAC Name* |
(1aR,4R,4aR,7R,7aS,7bS)-1,1,4,7-tetramethyl-2,3,4a,5,6,7,7a,7b-octahydro-1aH-cyclopropa[e]azulen-4-ol
|
|
| SMILES |
C[C@@H]1CC[C@@H]2[C@@H]1[C@H]3[C@H](C3(C)C)CC[C@@]2(C)O
|
|
| InChI |
InChI=1S/C15H26O/c1-9-5-6-10-12(9)13-11(14(13,2)3)7-8-15(10,4)16/h9-13,16H,5-8H2,1-4H3/t9-,10-,11-,12-,13-,15-/m1/s1
|
|
| InChIKey |
AYXPYQRXGNDJFU-QTPLKFIXSA-N
|
|
| Synonyms |
(-)-Globulol; Globulol; 489-41-8; G66H9XM0JK; (1aR,4R,4aR,7R,7aS,7bS)-1,1,4,7-tetramethyl-2,3,4a,5,6,7,7a,7b-octahydro-1aH-cyclopropa[e]azulen-4-ol; UNII-G66H9XM0JK; NSC-152470; EINECS 207-696-7; NSC 152470; SCHEMBL60792; GLOBULOL, (-)-; CHEMBL2171207; CHEBI:167415; DTXSID801318741; 1H-CYCLOPROP(E)AZULEN-4-OL, DECAHYDRO-1,1,4,7-TETRAMETHYL-, (1AR-(1A.ALPHA.,4.ALPHA.,4A.ALPHA.,7.ALPHA.,7A.BETA.,7B.ALPHA.))-; ZINC5528095; MFCD00042615; (1AR-(1aalpha,4alpha,4aalpha,7alpha,7abeta,7balpha))-decahydro-1,1,4,7-tetramethyl-1H-cycloprop(e)azulen-4-ol; (-)-Globulol, >=98.5% (sum of enantiomers, GC); Q27278826; (1AR-(1A.ALPHA.,4.ALPHA.,4A.ALPHA.,7.ALPHA.,7A.BETA.,7B.ALPHA.))-DECAHYDRO-1,1,4,7-TETRAMETHYL-1H-CYCLOPROP(E)AZULEN-4-OL; 1H-CYCLOPROP(E)AZULEN-4-OL, DECAHYDRO-1,1,4,7-TETRAMETHYL-, (1AR,4R,4AR,7R,7AS,7BS)-
|
|
| CAS | 489-41-8 | |
| PubChem CID | 12304985 | |
| ChEMBL ID | CHEMBL2171207 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 222.37 | ALogp: | 3.7 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 20.2 | Aromatic Rings: | 3 |
| Heavy Atoms: | 16 | QED Weighted: | 0.651 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.563 | MDCK Permeability: | 0.00003360 |
| Pgp-inhibitor: | 0.011 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.554 |
| 30% Bioavailability (F30%): | 0.469 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.131 | Plasma Protein Binding (PPB): | 96.12% |
| Volume Distribution (VD): | 1.286 | Fu: | 3.42% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.567 | CYP1A2-substrate: | 0.564 |
| CYP2C19-inhibitor: | 0.226 | CYP2C19-substrate: | 0.917 |
| CYP2C9-inhibitor: | 0.256 | CYP2C9-substrate: | 0.467 |
| CYP2D6-inhibitor: | 0.016 | CYP2D6-substrate: | 0.447 |
| CYP3A4-inhibitor: | 0.102 | CYP3A4-substrate: | 0.465 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 8.036 | Half-life (T1/2): | 0.44 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.09 | Human Hepatotoxicity (H-HT): | 0.109 |
| Drug-inuced Liver Injury (DILI): | 0.191 | AMES Toxicity: | 0.003 |
| Rat Oral Acute Toxicity: | 0.124 | Maximum Recommended Daily Dose: | 0.017 |
| Skin Sensitization: | 0.417 | Carcinogencity: | 0.033 |
| Eye Corrosion: | 0.985 | Eye Irritation: | 0.864 |
| Respiratory Toxicity: | 0.937 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC003089 | 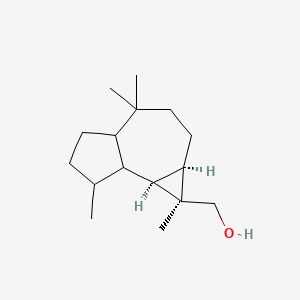 |
0.618 | D0N6FH |  |
0.375 | ||
| ENC001192 | 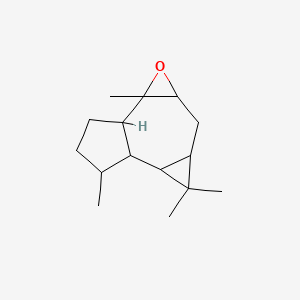 |
0.582 | D0S3WH | 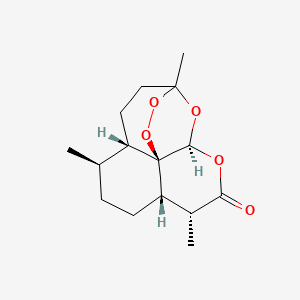 |
0.356 | ||
| ENC003084 | 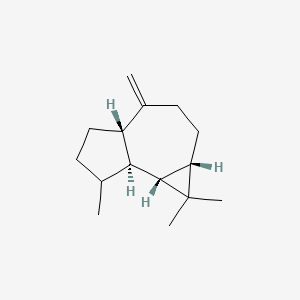 |
0.527 | D0Y5ZA | 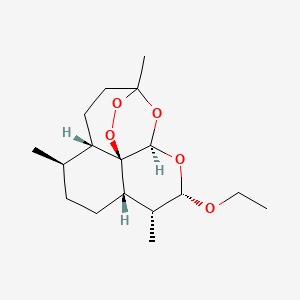 |
0.346 | ||
| ENC002256 | 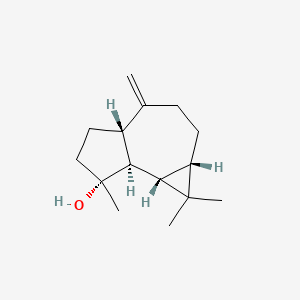 |
0.483 | D0U3GL | 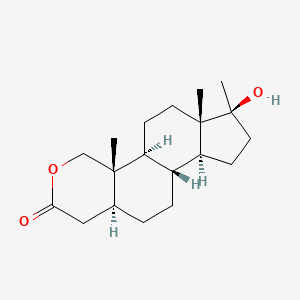 |
0.316 | ||
| ENC001196 | 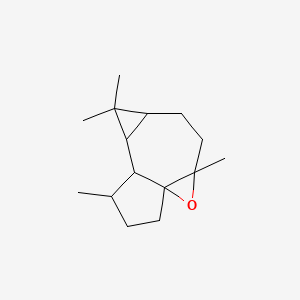 |
0.475 | D04DJN | 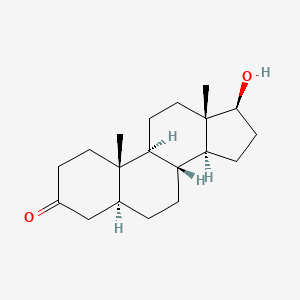 |
0.308 | ||
| ENC003088 | 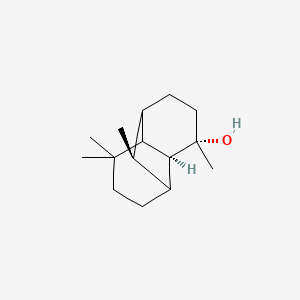 |
0.458 | D0B4RU | 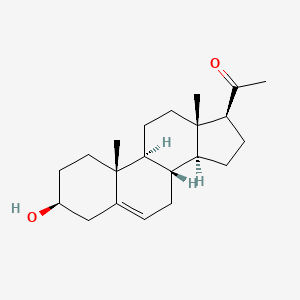 |
0.305 | ||
| ENC003050 | 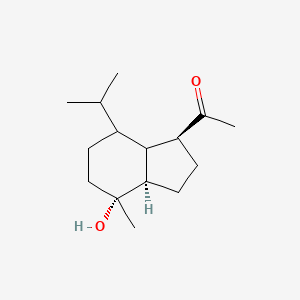 |
0.443 | D00VZZ | 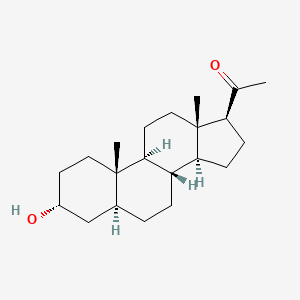 |
0.305 | ||
| ENC003624 | 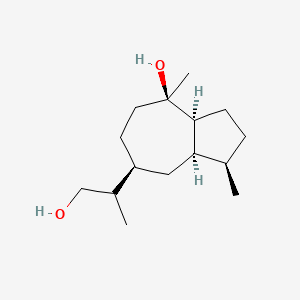 |
0.435 | D0Z1XD | 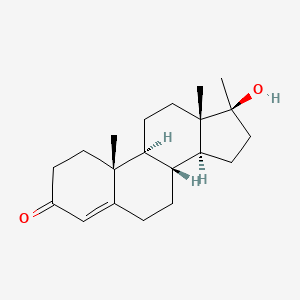 |
0.300 | ||
| ENC003125 | 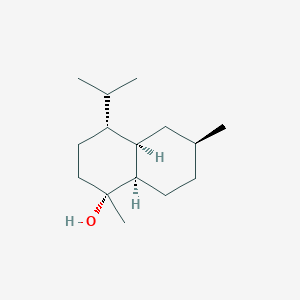 |
0.433 | D0Q6NZ | 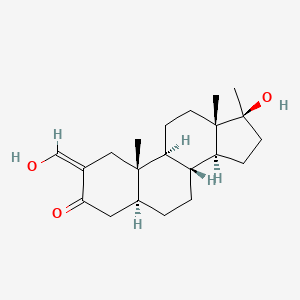 |
0.298 | ||
| ENC001321 | 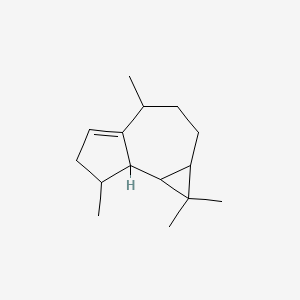 |
0.424 | D0D4JO |  |
0.297 | ||