NPs Basic Information

|
Name |
1,2-Dilinoleoylglycerol
|
| Molecular Formula | C39H68O5 | |
| IUPAC Name* |
[3-hydroxy-2-[(9Z,12Z)-octadeca-9,12-dienoyl]oxypropyl] (9Z,12Z)-octadeca-9,12-dienoate
|
|
| SMILES |
CCCCC/C=C\C/C=C\CCCCCCCC(=O)OCC(CO)OC(=O)CCCCCCC/C=C\C/C=C\CCCCC
|
|
| InChI |
InChI=1S/C39H68O5/c1-3-5-7-9-11-13-15-17-19-21-23-25-27-29-31-33-38(41)43-36-37(35-40)44-39(42)34-32-30-28-26-24-22-20-18-16-14-12-10-8-6-4-2/h11-14,17-20,37,40H,3-10,15-16,21-36H2,1-2H3/b13-11-,14-12-,19-17-,20-18-
|
|
| InChIKey |
MQGBAQLIFKSMEM-MAZCIEHSSA-N
|
|
| Synonyms |
1,2-Dilinoleoylglycerol; Glyceryl dilinoleate; 30606-27-0; 2442-62-8; glyceryl 1,2-dilinoleate; GF64S3WTPE; (9Z,12Z)-Octadeca-9,12-dienoic acid, diester with glycerol; UNII-GF64S3WTPE; dilinolein; EINECS 219-476-8; EINECS 250-259-0; Glycerol 1,2-dilinolate; DILINOLEIN (9c,12c); LINOLEIN, 1,2-DI-; SCHEMBL3181278; CHEBI:183969; DTXSID701021208; (+/-)-1,2-DILINOLEIN; [3-hydroxy-2-[(9Z,12Z)-octadeca-9,12-dienoyl]oxypropyl] (9Z,12Z)-octadeca-9,12-dienoate; J-015522; Q27279076; 9,12-Octadecadienoic acid, diester with 1,2,3-propanetriol; 1-(Hydroxymethyl)ethane-1,2-diyl bis((9Z,12Z)-octadeca-9,12-dienoate); 9,12-OCTADECADIENOIC ACID (9Z,12Z)-, 1-(HYDROXYMETHYL)-1,2-ETHANEDIYL ESTER; 9,12-OCTADECADIENOIC ACID (Z,Z)-, 1-(HYDROXYMETHYL)-1,2-ETHANEDIYL ESTER; 9,12-OCTADECADIENOIC ACID (9Z,12Z)-, 1,1'-(1-(HYDROXYMETHYL)-1,2-ETHANEDIYL) ESTER
|
|
| CAS | 2442-62-8 | |
| PubChem CID | 6438297 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 617.0 | ALogp: | 13.0 |
| HBD: | 1 | HBA: | 5 |
| Rotatable Bonds: | 34 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 72.8 | Aromatic Rings: | 0 |
| Heavy Atoms: | 44 | QED Weighted: | 0.037 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.105 | MDCK Permeability: | 0.00007090 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.266 | 20% Bioavailability (F20%): | 1 |
| 30% Bioavailability (F30%): | 1 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.005 | Plasma Protein Binding (PPB): | 101.37% |
| Volume Distribution (VD): | 3.071 | Fu: | 0.58% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.046 | CYP1A2-substrate: | 0.179 |
| CYP2C19-inhibitor: | 0.1 | CYP2C19-substrate: | 0.042 |
| CYP2C9-inhibitor: | 0.072 | CYP2C9-substrate: | 0.978 |
| CYP2D6-inhibitor: | 0.77 | CYP2D6-substrate: | 0.901 |
| CYP3A4-inhibitor: | 0.794 | CYP3A4-substrate: | 0.087 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 3.713 | Half-life (T1/2): | 0.94 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.315 | Human Hepatotoxicity (H-HT): | 0.234 |
| Drug-inuced Liver Injury (DILI): | 0.017 | AMES Toxicity: | 0.403 |
| Rat Oral Acute Toxicity: | 0.008 | Maximum Recommended Daily Dose: | 0.233 |
| Skin Sensitization: | 0.984 | Carcinogencity: | 0.111 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.019 |
| Respiratory Toxicity: | 0.509 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001766 | 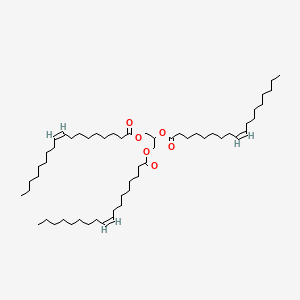 |
0.550 | D0O1TC | 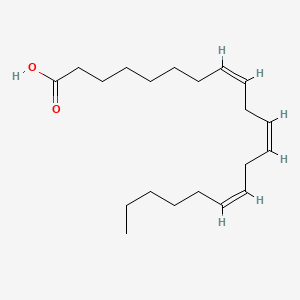 |
0.450 | ||
| ENC003072 | 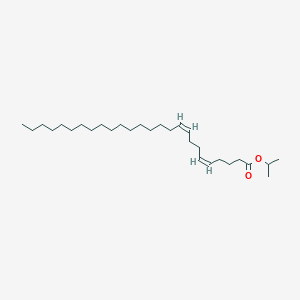 |
0.532 | D00MLW | 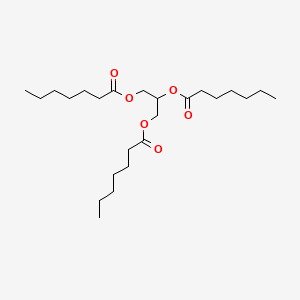 |
0.445 | ||
| ENC00491113 |  |
0.509 | D0Z1QC | 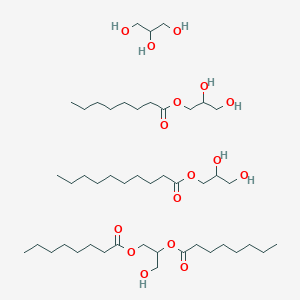 |
0.414 | ||
| ENC001628 | 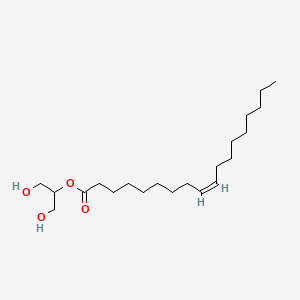 |
0.500 | D0O1PH |  |
0.409 | ||
| ENC001715 | 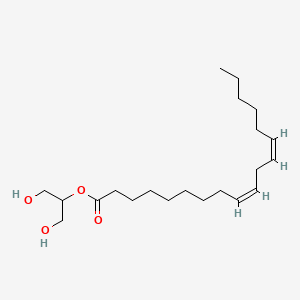 |
0.500 | D0UE9X | 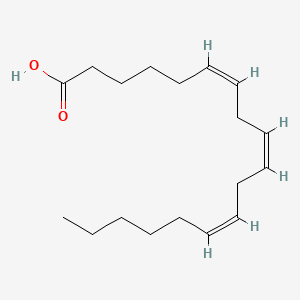 |
0.405 | ||
| ENC001845 |  |
0.496 | D0OR6A |  |
0.351 | ||
| ENC001700 |  |
0.489 | D0G7WY |  |
0.350 | ||
| ENC001678 | 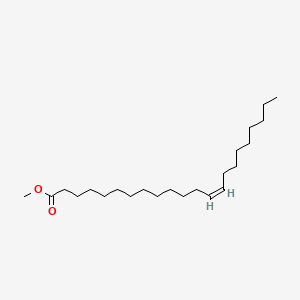 |
0.474 | D0G2MW | 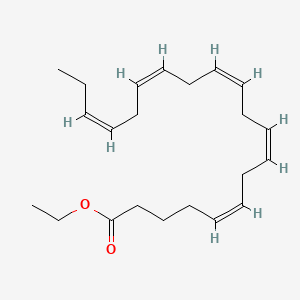 |
0.342 | ||
| ENC001643 | 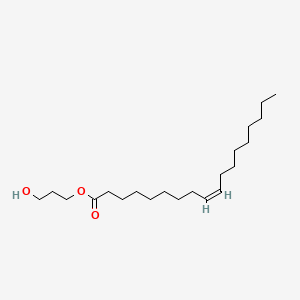 |
0.474 | D00AOJ | 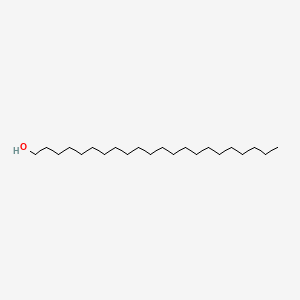 |
0.329 | ||
| ENC001711 |  |
0.473 | D0PS6X |  |
0.328 | ||