NPs Basic Information
|
Name |
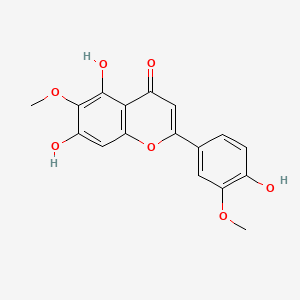
Jaceosidin
|
| Molecular Formula | C17H14O7 | |
| IUPAC Name* |
5,7-dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-6-methoxychromen-4-one
|
|
| SMILES |
COC1=C(C=CC(=C1)C2=CC(=O)C3=C(O2)C=C(C(=C3O)OC)O)O
|
|
| InChI |
InChI=1S/C17H14O7/c1-22-13-5-8(3-4-9(13)18)12-6-10(19)15-14(24-12)7-11(20)17(23-2)16(15)21/h3-7,18,20-21H,1-2H3
|
|
| InChIKey |
GLAAQZFBFGEBPS-UHFFFAOYSA-N
|
|
| Synonyms |
Jaceosidin; 18085-97-7; 5,7-Dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-6-methoxy-4H-chromen-4-one; 4',5,7-Trihydroxy-3',6-dimethoxyflavone; CHEBI:66103; 5U4Y68G678; 5,7-dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-6-methoxychromen-4-one; 4H-1-Benzopyran-4-one, 5,7-dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-6-methoxy-; 5,7-dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-6-methoxy-4H-1-Benzopyran-4-one; JACEOSIDINE; JASEOCIDIN; CHEMBL487601; SCHEMBL3960077; UNII-5U4Y68G678; BDBM84984; DTXSID00171022; BCP10203; HY-N0831; LMPK12111235; MFCD01081948; SC3961; ZB1886; ZINC14779854; AKOS015917411; AC-7927; 4,5,7-Trihydroxy-3,6-dimethoxyflavone; AS-73451; CS-0009862; FT-0686647; S0931; 085T977; Q-100215; FLAVONE, 4',5,7-TRIHYDROXY-3',6-DIMETHOXY-; Q27134618; B0005-464792; 5,7-Dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-6-methoxy-4H-chromen-4-one #
|
|
| CAS | 18085-97-7 | |
| PubChem CID | 5379096 | |
| ChEMBL ID | CHEMBL487601 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 330.29 | ALogp: | 1.7 |
| HBD: | 3 | HBA: | 7 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 105.0 | Aromatic Rings: | 3 |
| Heavy Atoms: | 24 | QED Weighted: | 0.675 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.96 | MDCK Permeability: | 0.00001230 |
| Pgp-inhibitor: | 0.42 | Pgp-substrate: | 0.383 |
| Human Intestinal Absorption (HIA): | 0.064 | 20% Bioavailability (F20%): | 0.015 |
| 30% Bioavailability (F30%): | 0.859 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.003 | Plasma Protein Binding (PPB): | 93.45% |
| Volume Distribution (VD): | 0.628 | Fu: | 13.43% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.95 | CYP1A2-substrate: | 0.952 |
| CYP2C19-inhibitor: | 0.24 | CYP2C19-substrate: | 0.065 |
| CYP2C9-inhibitor: | 0.726 | CYP2C9-substrate: | 0.838 |
| CYP2D6-inhibitor: | 0.477 | CYP2D6-substrate: | 0.769 |
| CYP3A4-inhibitor: | 0.45 | CYP3A4-substrate: | 0.186 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.742 | Half-life (T1/2): | 0.834 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.09 | Human Hepatotoxicity (H-HT): | 0.062 |
| Drug-inuced Liver Injury (DILI): | 0.898 | AMES Toxicity: | 0.471 |
| Rat Oral Acute Toxicity: | 0.046 | Maximum Recommended Daily Dose: | 0.342 |
| Skin Sensitization: | 0.871 | Carcinogencity: | 0.065 |
| Eye Corrosion: | 0.004 | Eye Irritation: | 0.913 |
| Respiratory Toxicity: | 0.342 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001573 | 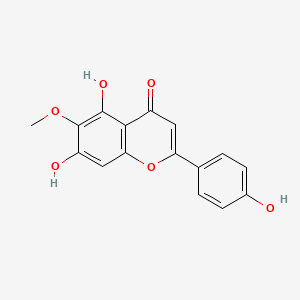 |
0.693 | D06GCK |  |
0.800 | ||
| ENC001534 | 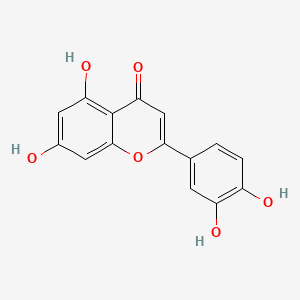 |
0.550 | D04AIT |  |
0.550 | ||
| ENC003106 | 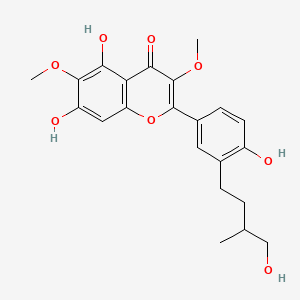 |
0.526 | D0K8KX | 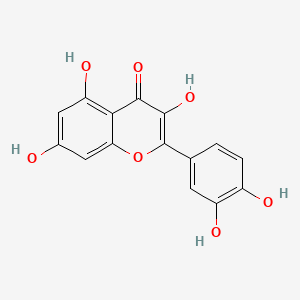 |
0.448 | ||
| ENC001772 | 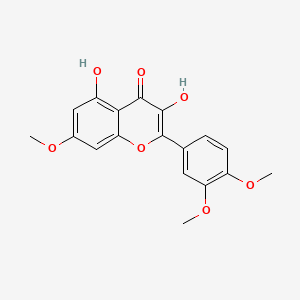 |
0.517 | D07MGA |  |
0.380 | ||
| ENC002471 | 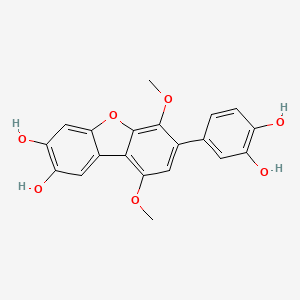 |
0.516 | D0G4KG | 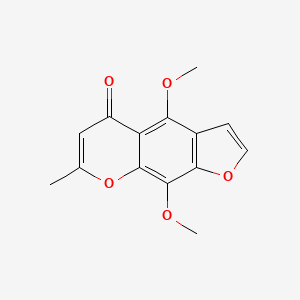 |
0.337 | ||
| ENC002475 | 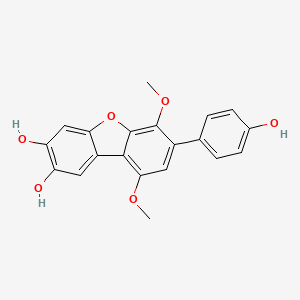 |
0.511 | D0TC7C | 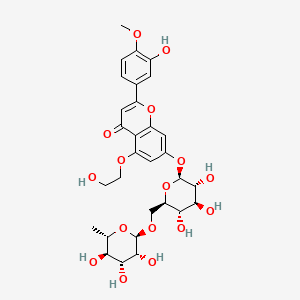 |
0.331 | ||
| ENC001533 | 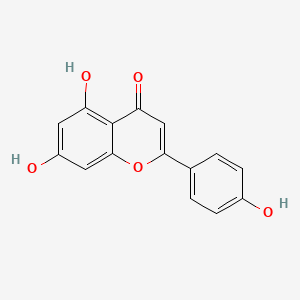 |
0.488 | D0AZ8C | 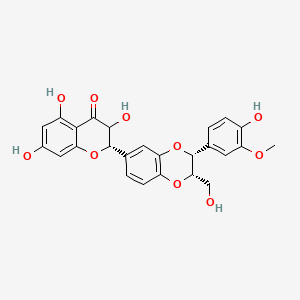 |
0.323 | ||
| ENC002853 |  |
0.473 | D0W8WB |  |
0.299 | ||
| ENC001631 |  |
0.465 | D02LZB |  |
0.294 | ||
| ENC002626 |  |
0.462 | D0Y7TS | 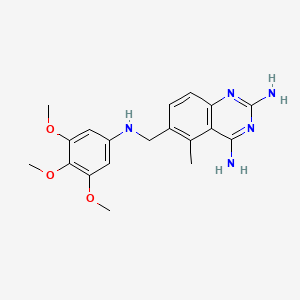 |
0.282 | ||