NPs Basic Information
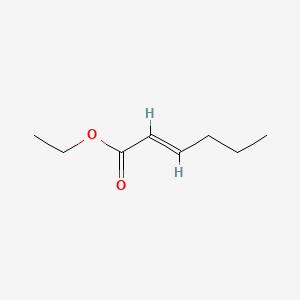
|
Name |
Ethyl trans-2-hexenoate
|
| Molecular Formula | C8H14O2 | |
| IUPAC Name* |
ethyl (E)-hex-2-enoate
|
|
| SMILES |
CCC/C=C/C(=O)OCC
|
|
| InChI |
InChI=1S/C8H14O2/c1-3-5-6-7-8(9)10-4-2/h6-7H,3-5H2,1-2H3/b7-6+
|
|
| InChIKey |
SJRXWMQZUAOMRJ-VOTSOKGWSA-N
|
|
| Synonyms |
27829-72-7; Ethyl hex-2-enoate; Ethyl trans-2-hexenoate; Ethyl 2-hexenoate; ethyl (E)-hex-2-enoate; 2-Hexenoic acid, ethyl ester; 2-Hexenoic acid, ethyl ester, (2E)-; 1552-67-6; Ethyl (E)-2-hexenoate; Ethyl 2E-hexenoate; (2E)-2-Hexenoic Acid Ethyl Ester; FEMA No. 3675; (E)-ethyl hex-2-enoate; 2-Hexenoic acid, ethyl ester, (E)-; ethyl (2E)-hex-2-enoate; OL4EUZ6C13; Ethyl ester of 2-hexenoic acid; trans-2-Hexenoic Acid Ethyl Ester; Ethyl (E)hex-2-enoate; UNII-OL4EUZ6C13; EINECS 216-296-1; EINECS 248-681-5; Ethyl E-hex-2-enoate; AI3-33376; Ethyl(E)-hex-2-enoate; SCHEMBL755909; (E)-ETHYL 2-HEXENOATE; CHEBI:87514; DTXSID30885403; (E)-2-Hexenoic acid ethyl ester; (E)-Hex-2-enoic acid ethyl ester; ALBB-032755; CBA82972; ZINC2384556; FEMA NO. 4613, E-; LMFA07010852; MFCD00036541; AKOS015903428; CS-W018283; ETHYL TRANS-2-HEXENOATE [FHFI]; LS-13479; DB-003641; E0787; EN300-7399033; A876859; J-016890; (R)-tert-butyl 3-((S)-4-benzyl-2-oxooxazolidine-3- carbonyl)hexanoate
|
|
| CAS | 27829-72-7 | |
| PubChem CID | 5364778 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 142.2 | ALogp: | 2.3 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 5 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 26.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 10 | QED Weighted: | 0.445 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.223 | MDCK Permeability: | 0.00004550 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.263 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.891 | Plasma Protein Binding (PPB): | 63.96% |
| Volume Distribution (VD): | 0.903 | Fu: | 33.79% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.949 | CYP1A2-substrate: | 0.226 |
| CYP2C19-inhibitor: | 0.51 | CYP2C19-substrate: | 0.517 |
| CYP2C9-inhibitor: | 0.096 | CYP2C9-substrate: | 0.599 |
| CYP2D6-inhibitor: | 0.023 | CYP2D6-substrate: | 0.244 |
| CYP3A4-inhibitor: | 0.064 | CYP3A4-substrate: | 0.239 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.909 | Half-life (T1/2): | 0.857 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.02 | Human Hepatotoxicity (H-HT): | 0.01 |
| Drug-inuced Liver Injury (DILI): | 0.044 | AMES Toxicity: | 0.016 |
| Rat Oral Acute Toxicity: | 0.019 | Maximum Recommended Daily Dose: | 0.014 |
| Skin Sensitization: | 0.899 | Carcinogencity: | 0.735 |
| Eye Corrosion: | 0.99 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.093 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000415 | 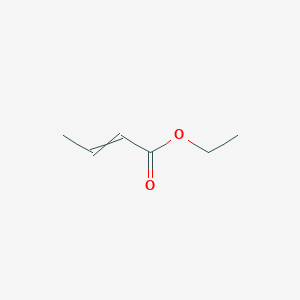 |
0.548 | D0Y3KG | 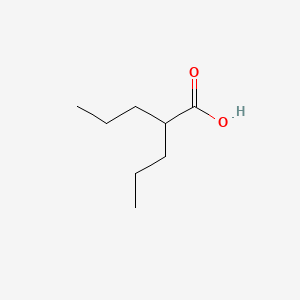 |
0.262 | ||
| ENC001586 | 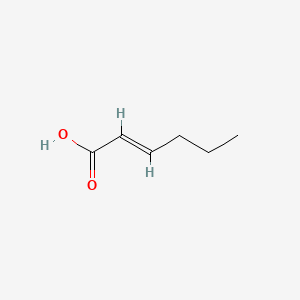 |
0.500 | D0A7MY | 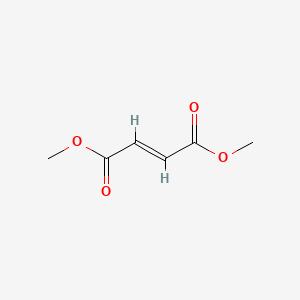 |
0.233 | ||
| ENC001463 | 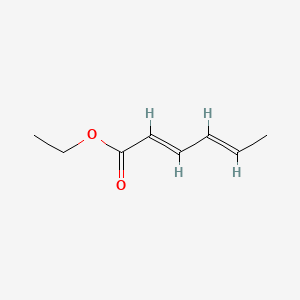 |
0.459 | D0G2MW | 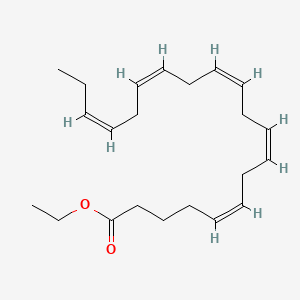 |
0.231 | ||
| ENC000226 | 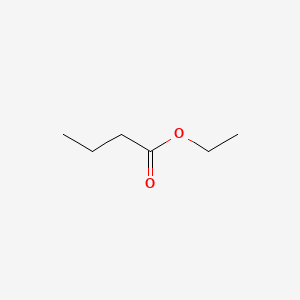 |
0.455 | D02CKX | 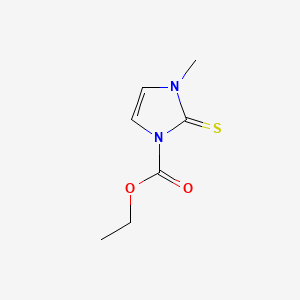 |
0.229 | ||
| ENC001642 | 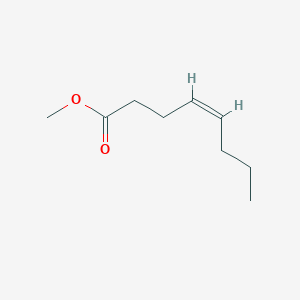 |
0.425 | D0Y4AW | 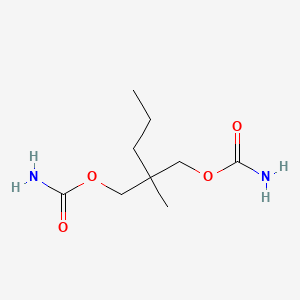 |
0.222 | ||
| ENC000371 | 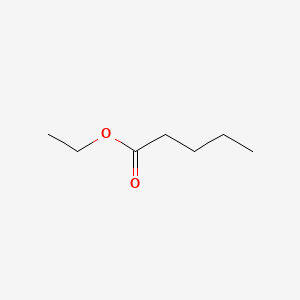 |
0.417 | D0K3LW | 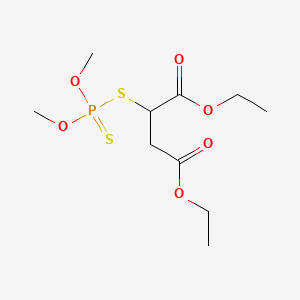 |
0.222 | ||
| ENC000224 | 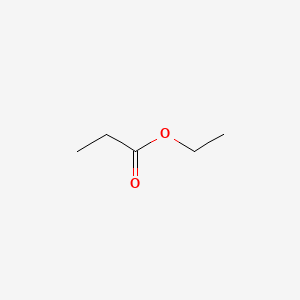 |
0.406 | D0Q9HF | 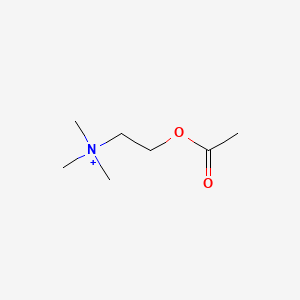 |
0.209 | ||
| ENC001668 | 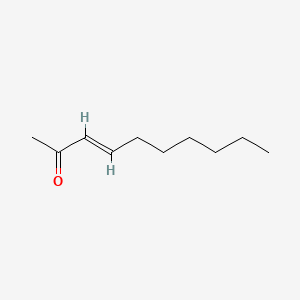 |
0.390 | D0U7BW | 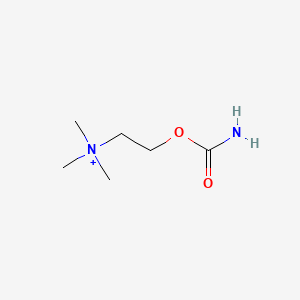 |
0.209 | ||
| ENC002790 | 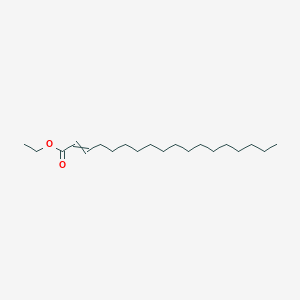 |
0.385 | D09CIQ | 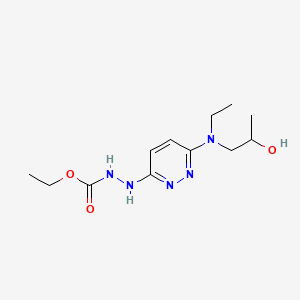 |
0.206 | ||
| ENC005738 | 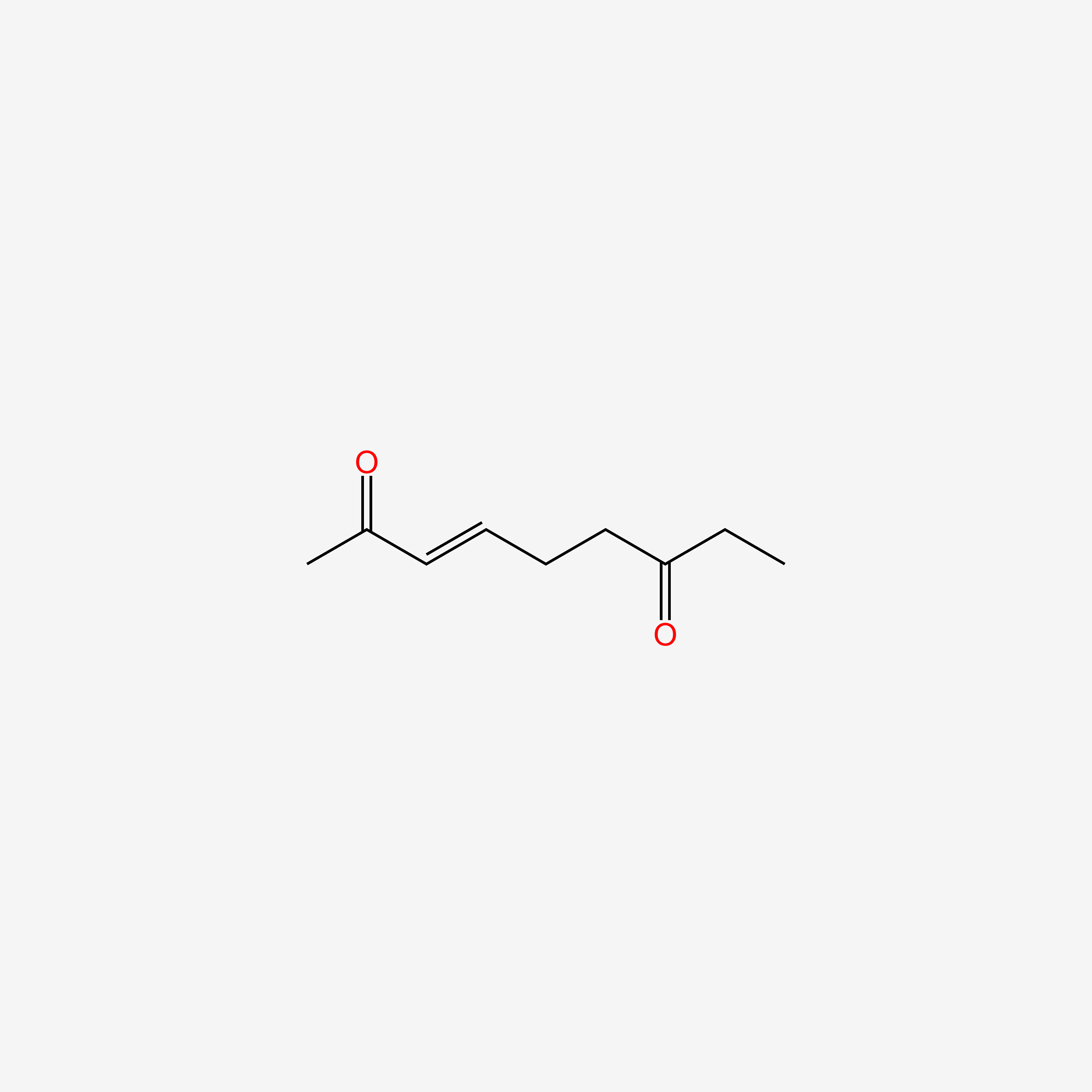 |
0.366 | D0ZK8H | 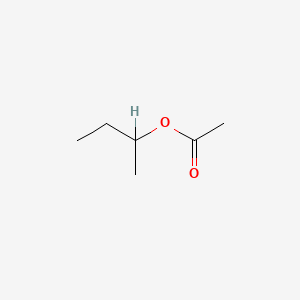 |
0.205 | ||