NPs Basic Information
|
Name |
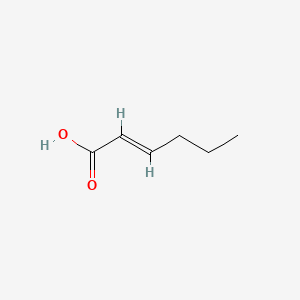
trans-2-Hexenoic acid
|
| Molecular Formula | C6H10O2 | |
| IUPAC Name* |
(E)-hex-2-enoic acid
|
|
| SMILES |
CCC/C=C/C(=O)O
|
|
| InChI |
InChI=1S/C6H10O2/c1-2-3-4-5-6(7)8/h4-5H,2-3H2,1H3,(H,7,8)/b5-4+
|
|
| InChIKey |
NIONDZDPPYHYKY-SNAWJCMRSA-N
|
|
| Synonyms |
trans-2-Hexenoic acid; 13419-69-7; (E)-hex-2-enoic acid; 2-HEXENOIC ACID; hex-2-enoic acid; trans-Hex-2-enoic acid; 1191-04-4; 2-Hexenoic acid, (2E)-; (E)-2-Hexenoic acid; Hexenoic acid; (2E)-2-Hexenoic acid; (2E)-hex-2-enoic acid; 2-Hexenoic acid, (E)-; 2E-hexenoic acid; Isohydrosorbic acid; (2E)-HEXENOIC ACID; FEMA No. 3169; beta-propyl acrylic acid; 2-Hexenoic acid, trans-; VQ24908VRU; CHEBI:87721; C6:1n-4; trans-2-hexenoicacid; 3-Propylacrylic acid; Propylacrylic acid, beta-; UNII-VQ24908VRU; UNII-BXY1L20879; hex-2-enoicacid; Hex-2-ensaeure; (E)-hexenoic acid; EINECS 214-727-8; EINECS 236-528-5; alpha.beta-Hexensaeure; beta-propylacrylic acid; alpha,beta-hexenoic acid; AI3-36119; SCHEMBL23614; SCHEMBL23615; trans-2-Hexenoic acid, 99%; QSPL 013; CHEMBL2252747; CHEBI:61206; NIONDZDPPYHYKY-SNAWJCMRSA-; DTXSID60884601; HEXENOIC ACID, TRANS 2-; BXY1L20879; ZINC1850698; LMFA01030008; MFCD00002705; AKOS000121254; TRANS-2-HEXENOIC ACID [FHFI]; C6:1, n-4; CS-W011247; trans-2-Hexenoic acid, >=98%, FG; 1289-40-3; BS-19492; CS-17340; DB-002108; H0383; EN300-21643; EN300-304058; A806747; Q-103481; Q27130883; Q27159869; Z104506794
|
|
| CAS | 13419-69-7 | |
| PubChem CID | 5282707 | |
| ChEMBL ID | CHEMBL2252747 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 114.14 | ALogp: | 1.6 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 37.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 8 | QED Weighted: | 0.569 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.614 | MDCK Permeability: | 0.00009290 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.001 |
| 30% Bioavailability (F30%): | 0.008 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.844 | Plasma Protein Binding (PPB): | 38.10% |
| Volume Distribution (VD): | 0.245 | Fu: | 40.17% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.022 | CYP1A2-substrate: | 0.116 |
| CYP2C19-inhibitor: | 0.022 | CYP2C19-substrate: | 0.087 |
| CYP2C9-inhibitor: | 0.005 | CYP2C9-substrate: | 0.924 |
| CYP2D6-inhibitor: | 0.007 | CYP2D6-substrate: | 0.195 |
| CYP3A4-inhibitor: | 0.011 | CYP3A4-substrate: | 0.051 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.438 | Half-life (T1/2): | 0.85 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.004 | Human Hepatotoxicity (H-HT): | 0.039 |
| Drug-inuced Liver Injury (DILI): | 0.025 | AMES Toxicity: | 0.01 |
| Rat Oral Acute Toxicity: | 0.029 | Maximum Recommended Daily Dose: | 0.017 |
| Skin Sensitization: | 0.339 | Carcinogencity: | 0.176 |
| Eye Corrosion: | 0.993 | Eye Irritation: | 0.995 |
| Respiratory Toxicity: | 0.134 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001587 |  |
0.543 | D0Y3KG |  |
0.270 | ||
| ENC001698 |  |
0.500 | D04CRL | 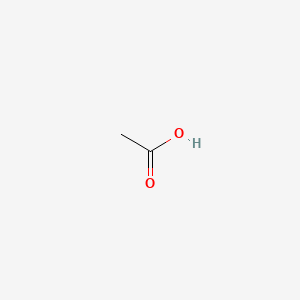 |
0.261 | ||
| ENC001588 | 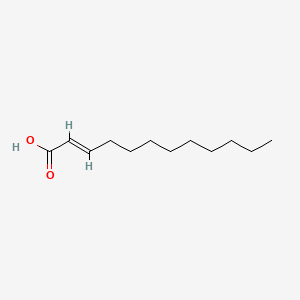 |
0.463 | D0N3NO | 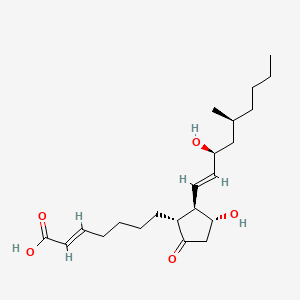 |
0.250 | ||
| ENC001585 | 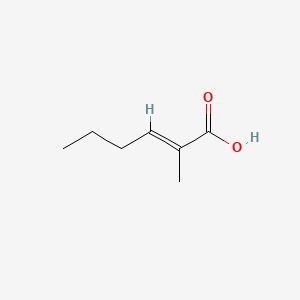 |
0.419 | D01ZJK | 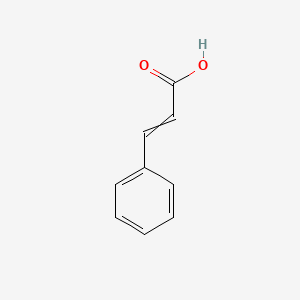 |
0.238 | ||
| ENC000018 | 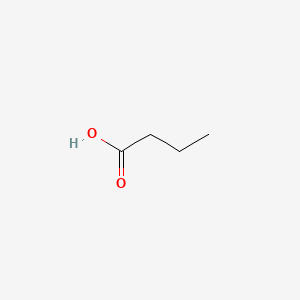 |
0.385 | D0M8AB | 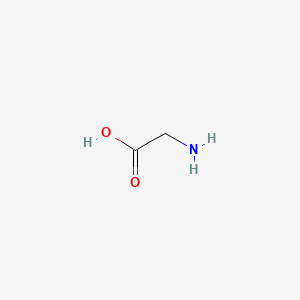 |
0.231 | ||
| ENC001668 | 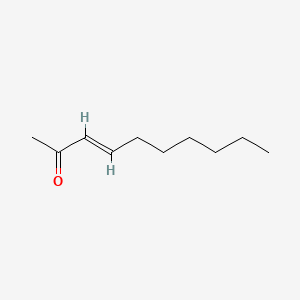 |
0.378 | D0R3QY |  |
0.222 | ||
| ENC001642 | 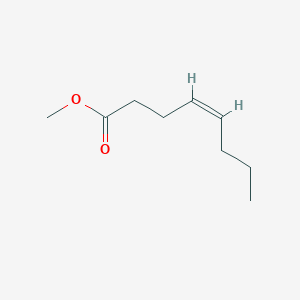 |
0.378 | D01OPV | 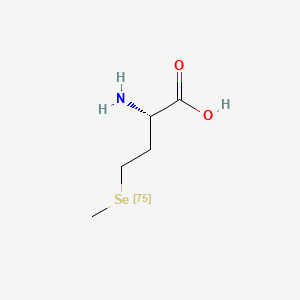 |
0.222 | ||
| ENC005738 | 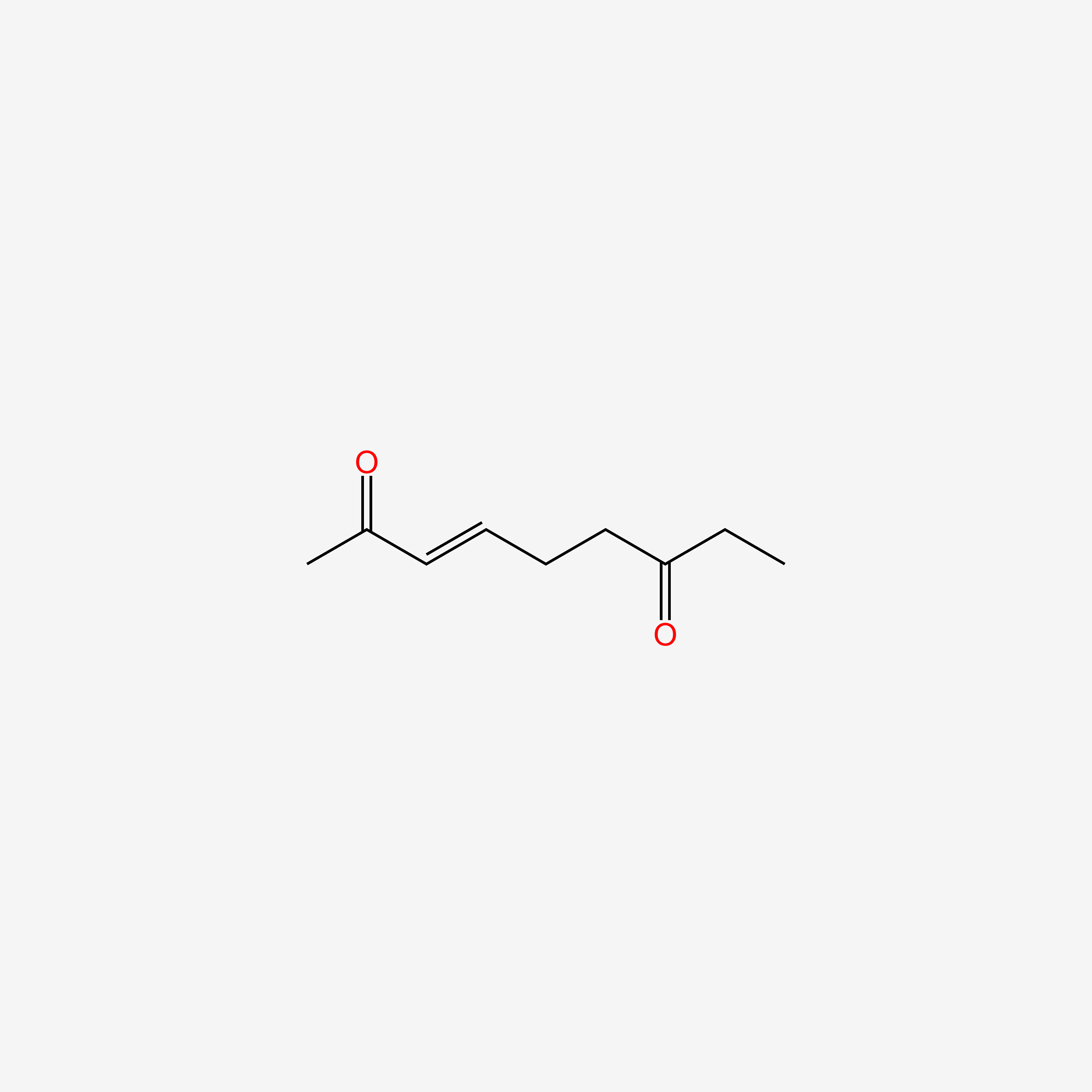 |
0.351 | D0UE9X | 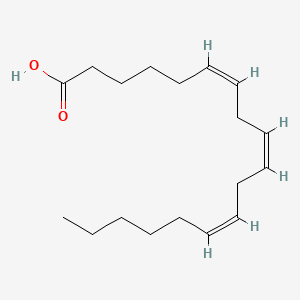 |
0.219 | ||
| ENC001725 | 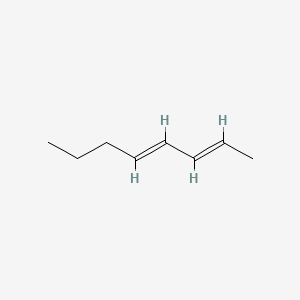 |
0.344 | D0EP8X | 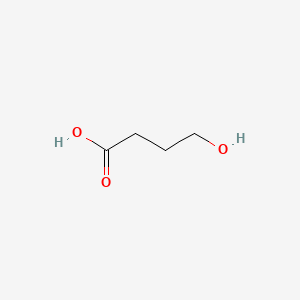 |
0.219 | ||
| ENC000058 | 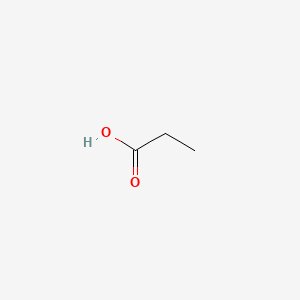 |
0.333 | D0V9EN | 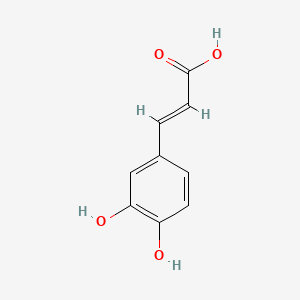 |
0.217 | ||