NPs Basic Information
|
Name |
(Z)-9-Hexadecenal
|
| Molecular Formula | C16H30O | |
| IUPAC Name* |
(Z)-hexadec-9-enal
|
|
| SMILES |
CCCCCC/C=C\CCCCCCCC=O
|
|
| InChI |
InChI=1S/C16H30O/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17/h7-8,16H,2-6,9-15H2,1H3/b8-7-
|
|
| InChIKey |
QFPVVMKZTVQDTL-FPLPWBNLSA-N
|
|
| Synonyms |
(Z)-Hexadec-9-enal; (Z)-9-Hexadecenal; 56219-04-6; cis-9-Hexadecenal; 9Z-Hexadecenal; 9-Hexadecenal, (9Z)-; Z-9-HEXADECENAL; 9-Hexadecenal, (Z)-; NXO9ML6I2E; (9Z)-9-Hexadecenal; EINECS 260-064-2; UNII-NXO9ML6I2E; (9Z)-9-Hexadecenal #; SCHEMBL556941; (9Z)-HEXADEC-9-ENAL; HEXADECENAL, (Z)-9-; DTXSID5041194; CIS-9-HEXADECENAL, 95; CHEBI:179683; 9Z-HEXADECEN/AL , 95 %; LMFA06000217; ZINC59725570; 9Z-Hexadecenal CAS 56219-04-6; Q27285093; 81K
|
|
| CAS | 56219-04-6 | |
| PubChem CID | 5364643 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 238.41 | ALogp: | 6.1 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 13 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 17 | QED Weighted: | 0.23 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.807 | MDCK Permeability: | 0.00002290 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.016 | 20% Bioavailability (F20%): | 0.998 |
| 30% Bioavailability (F30%): | 0.999 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.884 | Plasma Protein Binding (PPB): | 89.41% |
| Volume Distribution (VD): | 3.735 | Fu: | 2.94% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.362 | CYP1A2-substrate: | 0.269 |
| CYP2C19-inhibitor: | 0.394 | CYP2C19-substrate: | 0.076 |
| CYP2C9-inhibitor: | 0.252 | CYP2C9-substrate: | 0.932 |
| CYP2D6-inhibitor: | 0.446 | CYP2D6-substrate: | 0.706 |
| CYP3A4-inhibitor: | 0.412 | CYP3A4-substrate: | 0.076 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 3.858 | Half-life (T1/2): | 0.742 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.385 | Human Hepatotoxicity (H-HT): | 0.032 |
| Drug-inuced Liver Injury (DILI): | 0.033 | AMES Toxicity: | 0.168 |
| Rat Oral Acute Toxicity: | 0.021 | Maximum Recommended Daily Dose: | 0.043 |
| Skin Sensitization: | 0.976 | Carcinogencity: | 0.296 |
| Eye Corrosion: | 0.988 | Eye Irritation: | 0.989 |
| Respiratory Toxicity: | 0.939 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
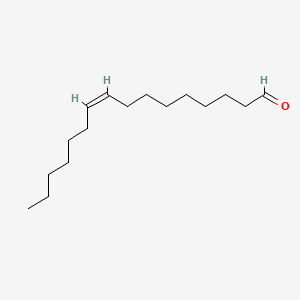
| ENC001686 | 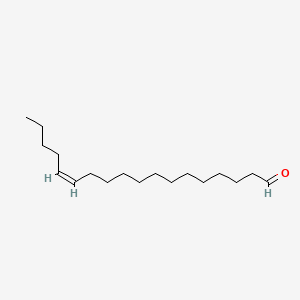 |
0.825 | D0O1PH |  |
0.575 | ||
| ENC001690 |  |
0.759 | D0O1TC |  |
0.534 | ||
| ENC001713 | 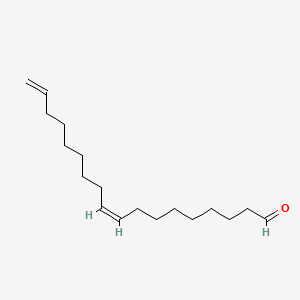 |
0.733 | D0OR6A | 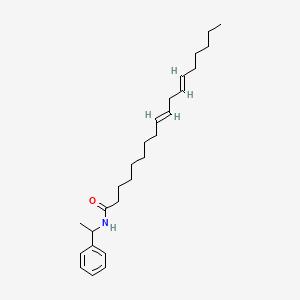 |
0.461 | ||
| ENC001099 | 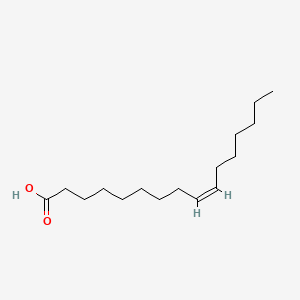 |
0.724 | D0UE9X | 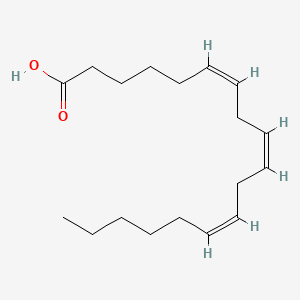 |
0.452 | ||
| ENC002845 | 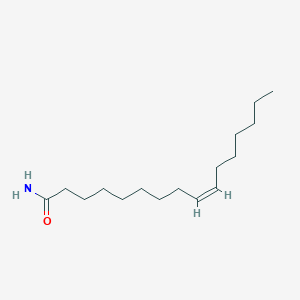 |
0.724 | D05ATI |  |
0.433 | ||
| ENC001589 | 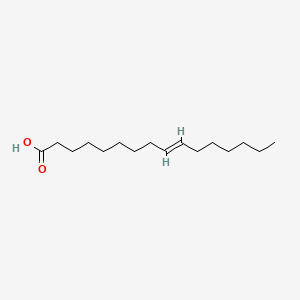 |
0.724 | D0Z5SM |  |
0.411 | ||
| ENC001435 | 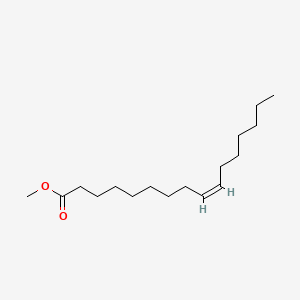 |
0.689 | D07ILQ |  |
0.380 | ||
| ENC000277 | 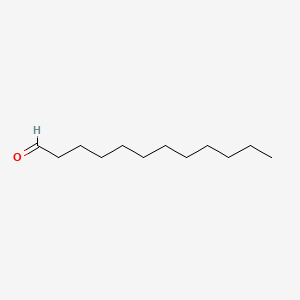 |
0.686 | D0Z5BC | 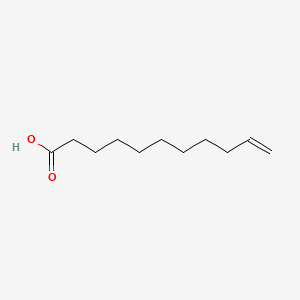 |
0.349 | ||
| ENC000607 | 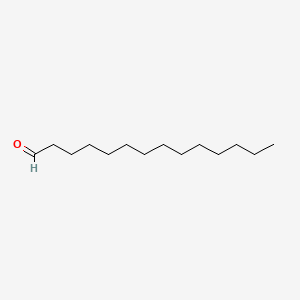 |
0.673 | D09SRR | 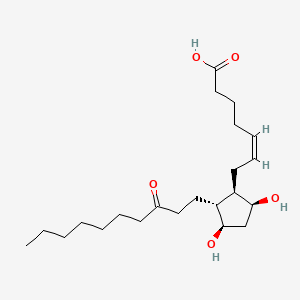 |
0.348 | ||
| ENC001666 | 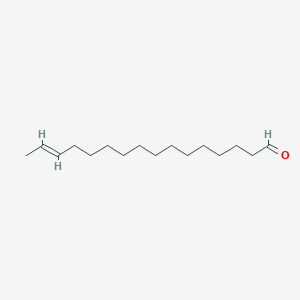 |
0.661 | D00AOJ |  |
0.333 | ||