NPs Basic Information
|
Name |
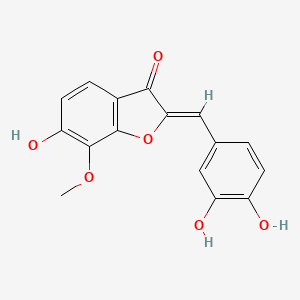
Leptosidin
|
| Molecular Formula | C16H12O6 | |
| IUPAC Name* |
(2Z)-2-[(3,4-dihydroxyphenyl)methylidene]-6-hydroxy-7-methoxy-1-benzofuran-3-one
|
|
| SMILES |
COC1=C(C=CC2=C1O/C(=C\C3=CC(=C(C=C3)O)O)/C2=O)O
|
|
| InChI |
InChI=1S/C16H12O6/c1-21-16-11(18)5-3-9-14(20)13(22-15(9)16)7-8-2-4-10(17)12(19)6-8/h2-7,17-19H,1H3/b13-7-
|
|
| InChIKey |
PFRGTMTYWMVLMU-QPEQYQDCSA-N
|
|
| Synonyms |
Leptosidin; 486-24-8; VE67J9PR82; 3(2H)-Benzofuranone, 2-(3,4-dihydroxybenzylidene)-6-hydroxy-7-methoxy-; C08651; (Z)-2-((3,4-Dihydroxyphenyl)methylene)-6-hydroxy-7-methoxy-3(2H)-benzofuranone; 3(2H)-Benzofuranone, 2-((3,4-dihydroxyphenyl)methylene)-6-hydroxy-7-methoxy-, (Z)-; AC1NQYAI; SureCN6804084; UNII-VE67J9PR82; CHEBI:6412; SCHEMBL6804084; CHEMBL3799890; DTXSID501028834; ZINC4642934; Q6528333; (2Z)-2-((3,4-DIHYDROXYPHENYL)METHYLENE)-6-HYDROXY-7-METHOXY-3(2H)-BENZOFURANONE; (2Z)-2-[(3,4-dihydroxyphenyl)methylene]-6-hydroxy-7-methoxy-benzofuran-3-one; (2Z)-2-[(3,4-dihydroxyphenyl)methylidene]-6-hydroxy-7-methoxy-1-benzofuran-3(2H)-one; 2-(3,4-Dihydroxybenzylidene)-6-hydroxy-7-methoxy-2,3-dihydrobenzofuran-3-one
|
|
| CAS | 486-24-8 | |
| PubChem CID | 5281257 | |
| ChEMBL ID | CHEMBL3799890 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 300.26 | ALogp: | 2.5 |
| HBD: | 3 | HBA: | 6 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 96.2 | Aromatic Rings: | 3 |
| Heavy Atoms: | 22 | QED Weighted: | 0.582 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.027 | MDCK Permeability: | 0.00001850 |
| Pgp-inhibitor: | 0.138 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.022 | 20% Bioavailability (F20%): | 0.489 |
| 30% Bioavailability (F30%): | 0.005 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.055 | Plasma Protein Binding (PPB): | 99.45% |
| Volume Distribution (VD): | 0.45 | Fu: | 2.08% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.947 | CYP1A2-substrate: | 0.306 |
| CYP2C19-inhibitor: | 0.171 | CYP2C19-substrate: | 0.057 |
| CYP2C9-inhibitor: | 0.606 | CYP2C9-substrate: | 0.437 |
| CYP2D6-inhibitor: | 0.216 | CYP2D6-substrate: | 0.241 |
| CYP3A4-inhibitor: | 0.495 | CYP3A4-substrate: | 0.198 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 8.579 | Half-life (T1/2): | 0.893 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.017 | Human Hepatotoxicity (H-HT): | 0.179 |
| Drug-inuced Liver Injury (DILI): | 0.546 | AMES Toxicity: | 0.61 |
| Rat Oral Acute Toxicity: | 0.961 | Maximum Recommended Daily Dose: | 0.916 |
| Skin Sensitization: | 0.927 | Carcinogencity: | 0.747 |
| Eye Corrosion: | 0.004 | Eye Irritation: | 0.937 |
| Respiratory Toxicity: | 0.279 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC003373 | 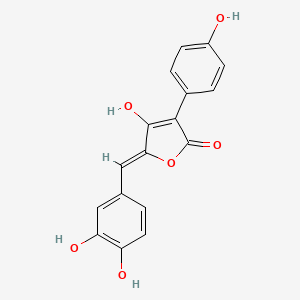 |
0.506 | D04AIT |  |
0.368 | ||
| ENC005410 | 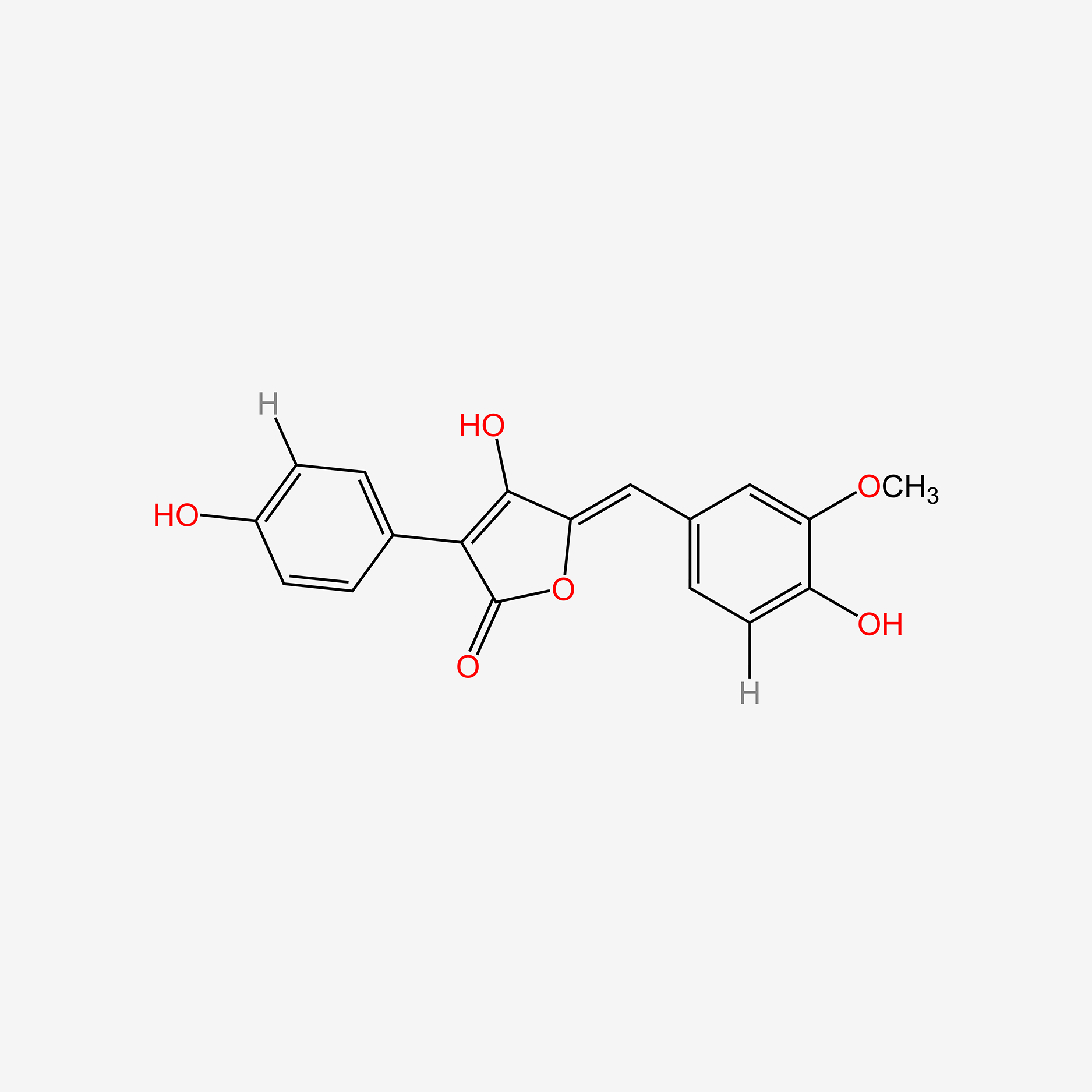 |
0.488 | D0K8KX | 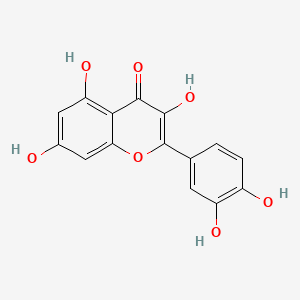 |
0.360 | ||
| ENC003492 | 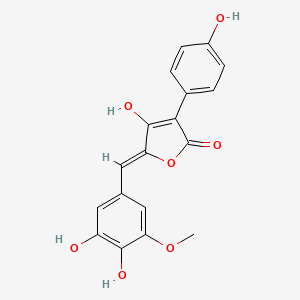 |
0.444 | D07MGA |  |
0.341 | ||
| ENC002823 | 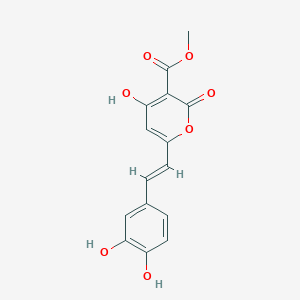 |
0.424 | D0V9EN | 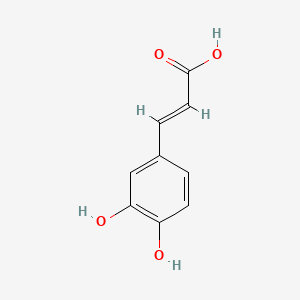 |
0.333 | ||
| ENC001998 | 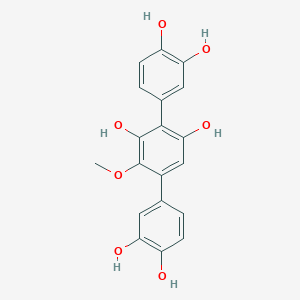 |
0.419 | D0U3YB | 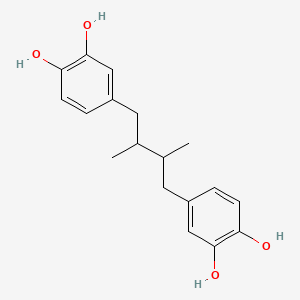 |
0.330 | ||
| ENC002756 | 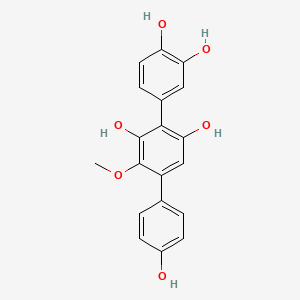 |
0.413 | D06GCK |  |
0.313 | ||
| ENC002125 | 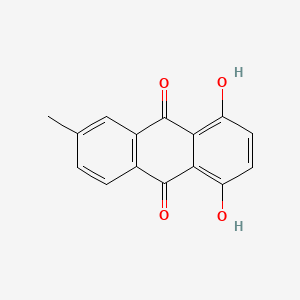 |
0.413 | D0E9CD | 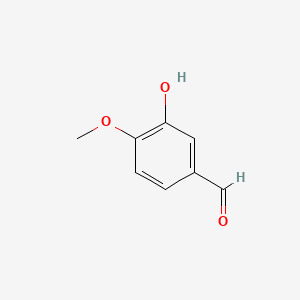 |
0.300 | ||
| ENC005040 | 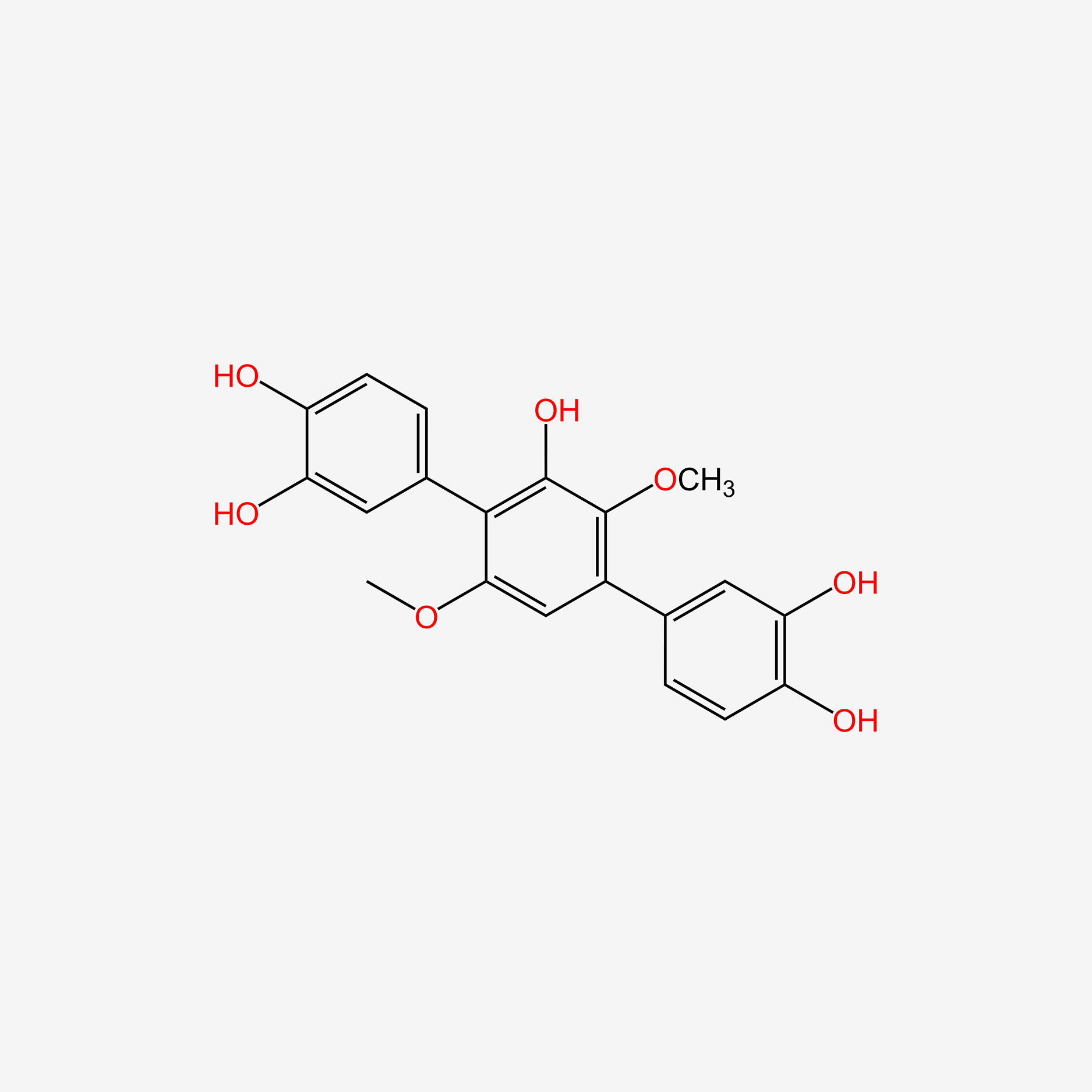 |
0.406 | D08SKH | 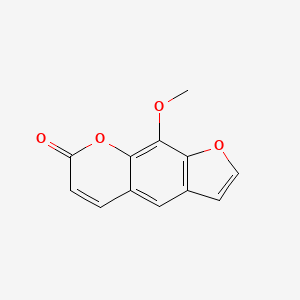 |
0.289 | ||
| ENC001573 | 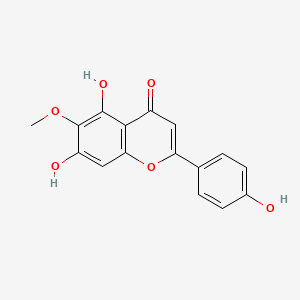 |
0.402 | D0AZ8C | 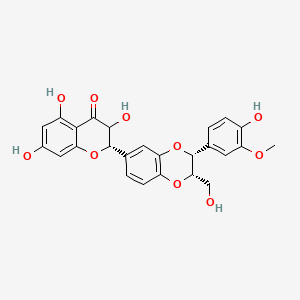 |
0.282 | ||
| ENC005411 | 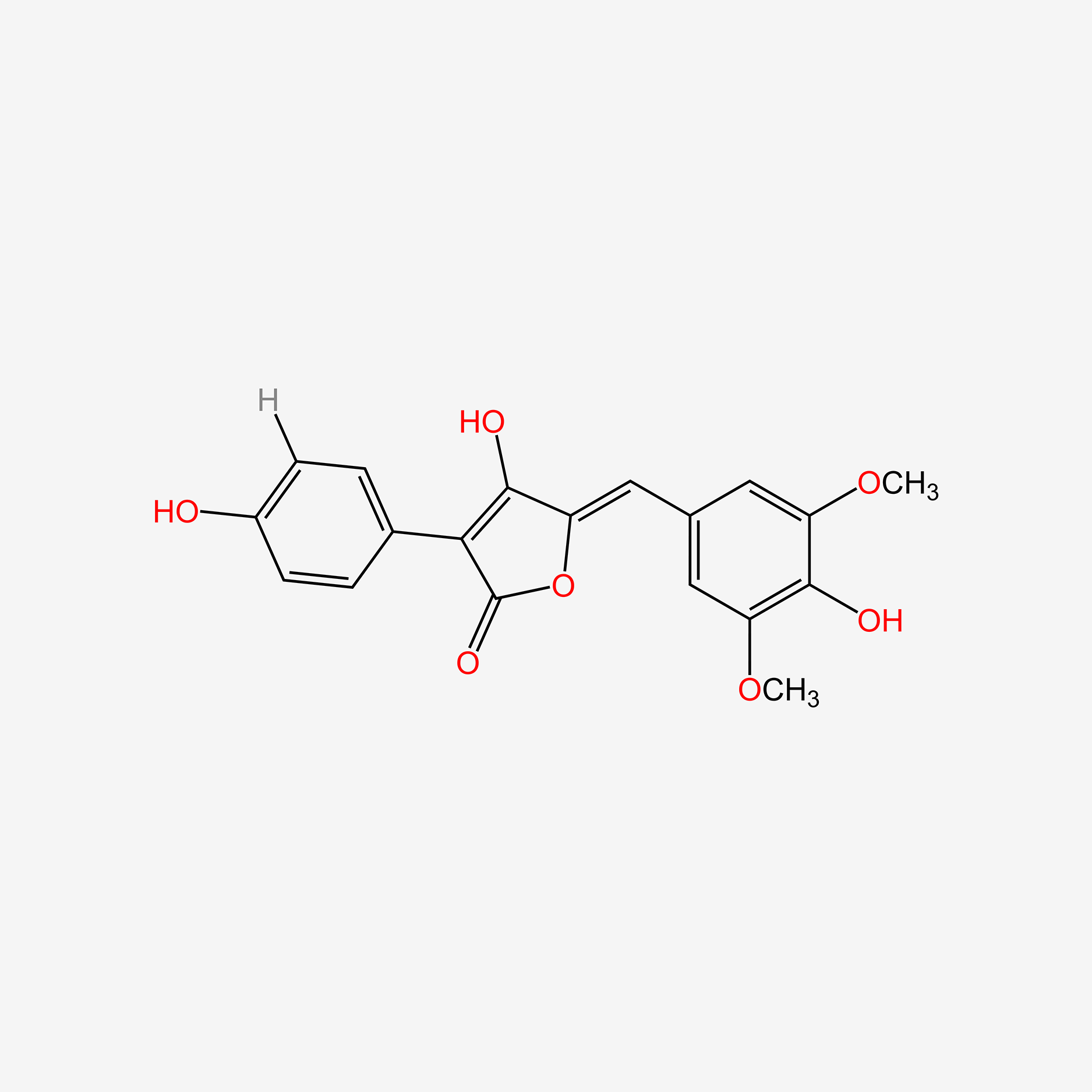 |
0.400 | D0J7RK | 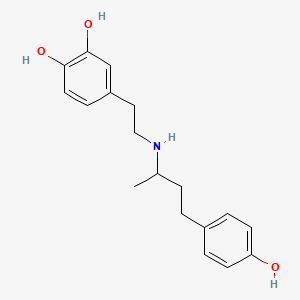 |
0.281 | ||