NPs Basic Information
|
Name |
Asterric acid
|
| Molecular Formula | C17H16O8 | |
| IUPAC Name* |
2-hydroxy-6-(4-hydroxy-2-methoxy-6-methoxycarbonylphenoxy)-4-methylbenzoic acid
|
|
| SMILES |
CC1=CC(=C(C(=C1)OC2=C(C=C(C=C2OC)O)C(=O)OC)C(=O)O)O
|
|
| InChI |
InChI=1S/C17H16O8/c1-8-4-11(19)14(16(20)21)12(5-8)25-15-10(17(22)24-3)6-9(18)7-13(15)23-2/h4-7,18-19H,1-3H3,(H,20,21)
|
|
| InChIKey |
XOKVHFNTYHPEHN-UHFFFAOYSA-N
|
|
| Synonyms |
Asterric acid; 577-64-0; J3Q23XL4KN; MLS000863583; CHEMBL469424; 2-hydroxy-6-(4-hydroxy-2-methoxy-6-methoxycarbonylphenoxy)-4-methylbenzoic acid; Benzoic acid, 2-(2-carboxy-3-hydroxy-5-methylphenoxy)-5-hydroxy-3-methoxy-, 1-methyl ester; SMR000440714; Asterric acid_120092; UNII-J3Q23XL4KN; MEGxm0_000244; SCHEMBL6046569; ZINC7778; ACon0_000930; ACon1_001529; BDBM50711; cid_3080568; TAN-1415A; XOKVHFNTYHPEHN-UHFFFAOYSA-; DTXSID60206405; HMS2270H17; NCGC00180411-01; Asterric acid, >=95% (LC/MS-ELSD); DB-127036; L007432; BRD-K20637966-001-01-0; Q15410258; Methyl 2-(2-carboxy-3-hydroxy-5-methylphenoxy)-5-hydroxy-3-methoxybenzoate; 2-(2-(methoxycarbonyl)-4-hydroxy-6-methoxyphenoxy)-6-hydroxy-4-methylbenzoic acid; 2-(2-carbomethoxy-4-hydroxy-6-methoxy-phenoxy)-6-hydroxy-4-methyl-benzoic acid; 2-(2-methoxy-6-methoxycarbonyl-4-oxidanyl-phenoxy)-4-methyl-6-oxidanyl-benzoic acid; 2-Hydroxy-6-(4-hydroxy-2-methoxy-6-(methoxycarbonyl)phenoxy)-4-methylbenzoic acid; 2-hydroxy-6-(4-hydroxy-2-methoxy-6-methoxycarbonyl-phenoxy)-4-meta??hyl-benzoic acid
|
|
| CAS | 577-64-0 | |
| PubChem CID | 3080568 | |
| ChEMBL ID | CHEMBL469424 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 348.3 | ALogp: | 3.0 |
| HBD: | 3 | HBA: | 8 |
| Rotatable Bonds: | 6 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 123.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 25 | QED Weighted: | 0.702 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.305 | MDCK Permeability: | 0.00000931 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.025 |
| Human Intestinal Absorption (HIA): | 0.191 | 20% Bioavailability (F20%): | 0.02 |
| 30% Bioavailability (F30%): | 0.029 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.064 | Plasma Protein Binding (PPB): | 94.77% |
| Volume Distribution (VD): | 0.444 | Fu: | 6.03% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.288 | CYP1A2-substrate: | 0.766 |
| CYP2C19-inhibitor: | 0.079 | CYP2C19-substrate: | 0.054 |
| CYP2C9-inhibitor: | 0.518 | CYP2C9-substrate: | 0.181 |
| CYP2D6-inhibitor: | 0.375 | CYP2D6-substrate: | 0.151 |
| CYP3A4-inhibitor: | 0.089 | CYP3A4-substrate: | 0.101 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.466 | Half-life (T1/2): | 0.894 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.022 | Human Hepatotoxicity (H-HT): | 0.287 |
| Drug-inuced Liver Injury (DILI): | 0.96 | AMES Toxicity: | 0.01 |
| Rat Oral Acute Toxicity: | 0.822 | Maximum Recommended Daily Dose: | 0.488 |
| Skin Sensitization: | 0.12 | Carcinogencity: | 0.017 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.291 |
| Respiratory Toxicity: | 0.736 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
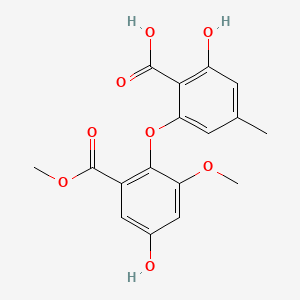
| ENC001522 | 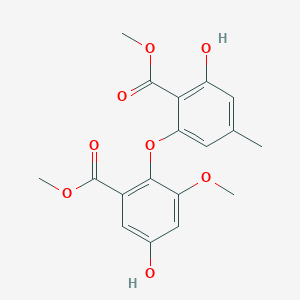 |
0.827 | D06GCK |  |
0.333 | ||
| ENC005416 | 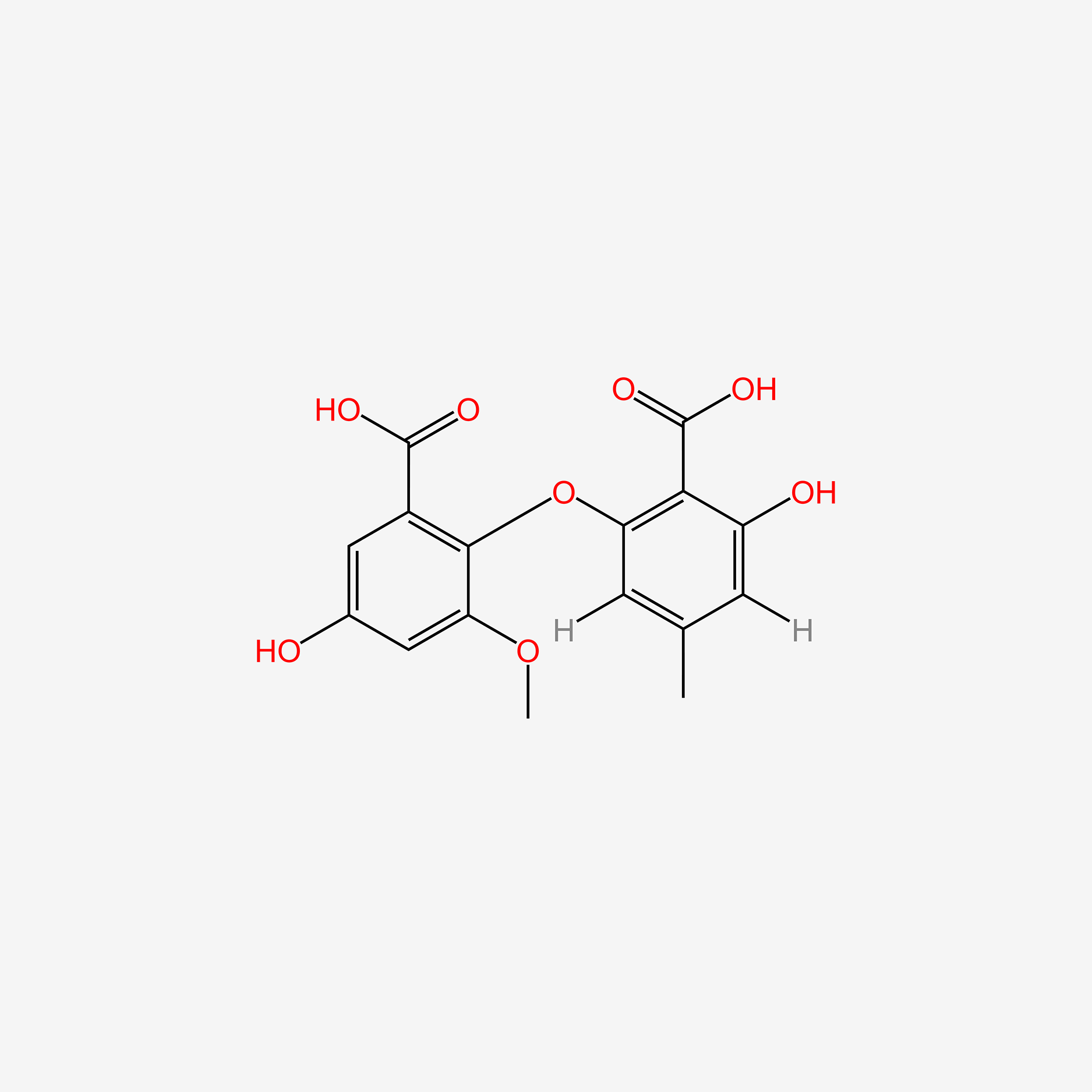 |
0.819 | D07MGA |  |
0.306 | ||
| ENC006015 | 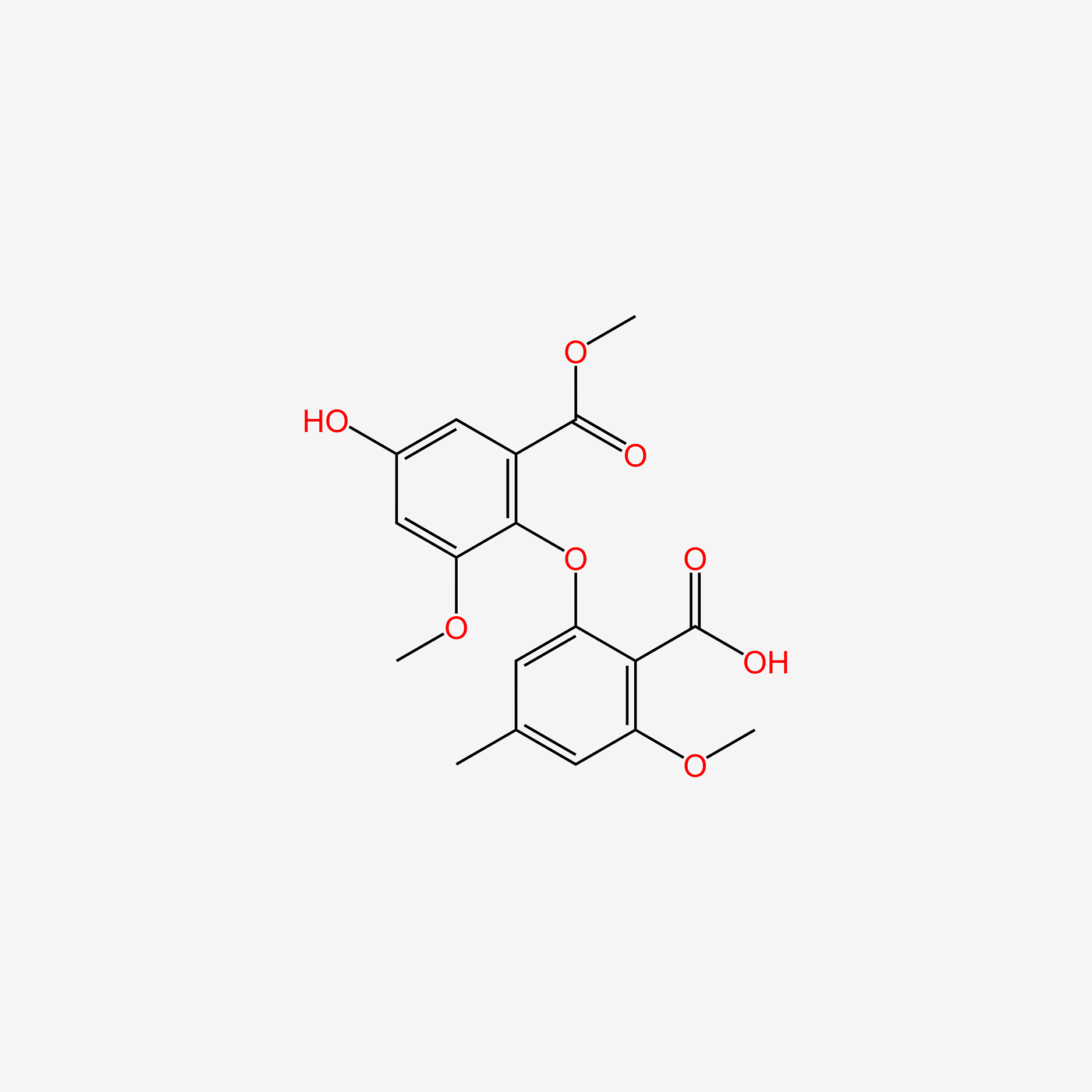 |
0.803 | D0E6OC | 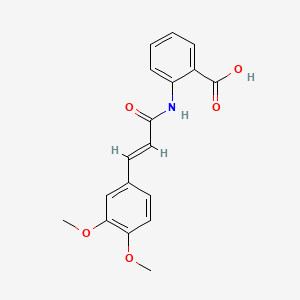 |
0.288 | ||
| ENC002526 | 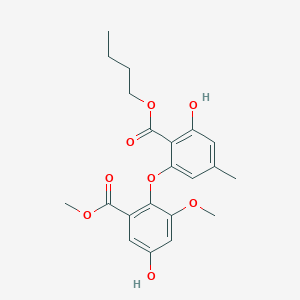 |
0.738 | D09DHY | 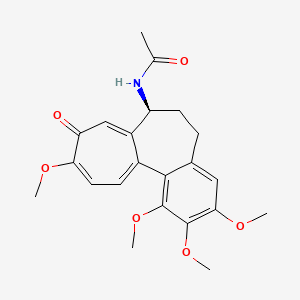 |
0.278 | ||
| ENC002381 | 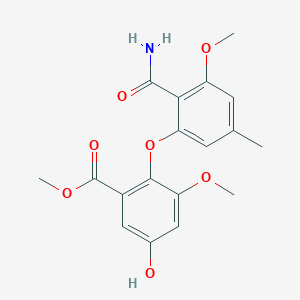 |
0.713 | D00KRE | 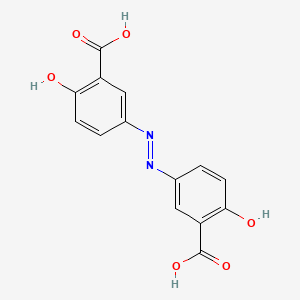 |
0.270 | ||
| ENC005978 | 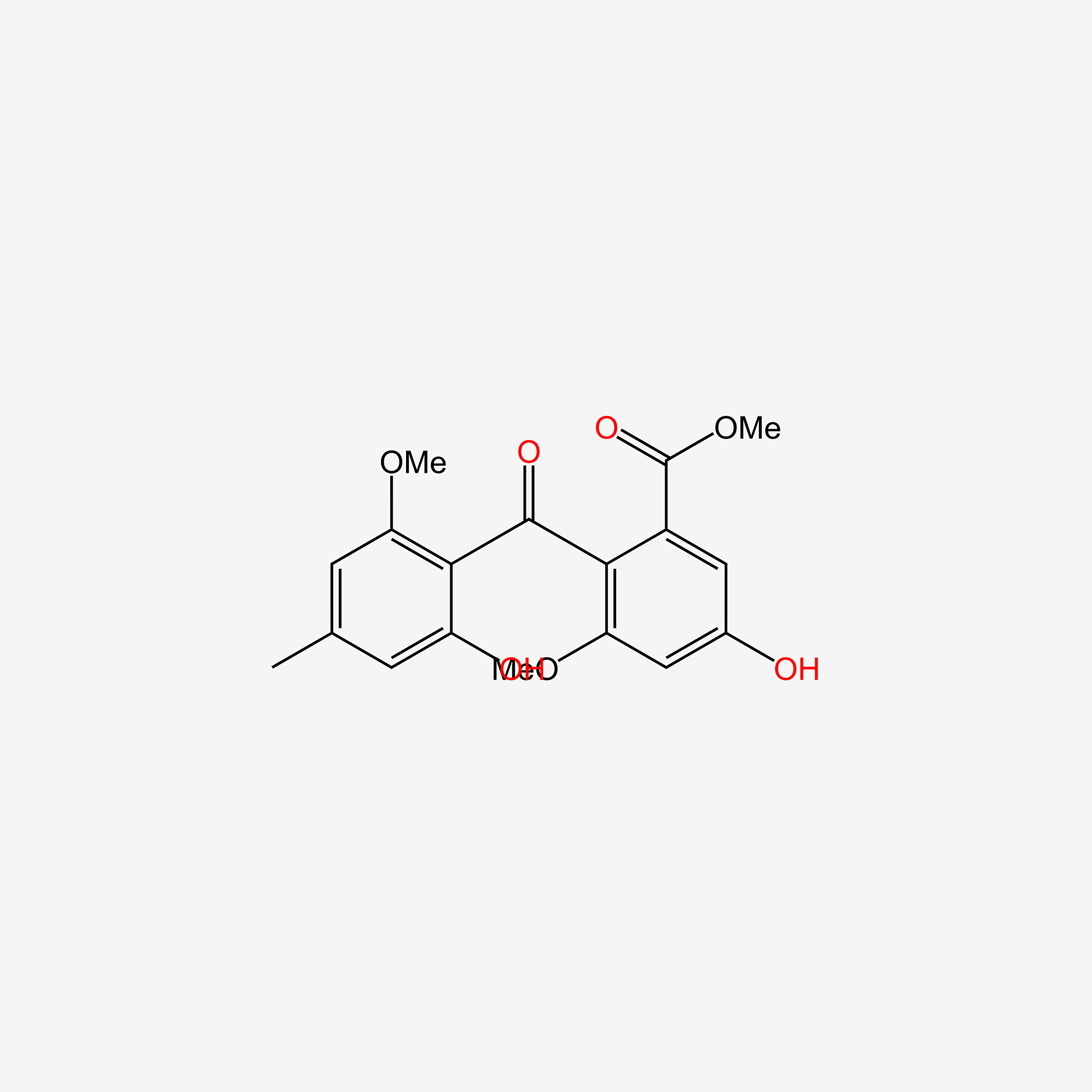 |
0.696 | D00WVW | 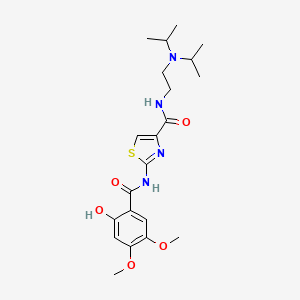 |
0.269 | ||
| ENC002468 | 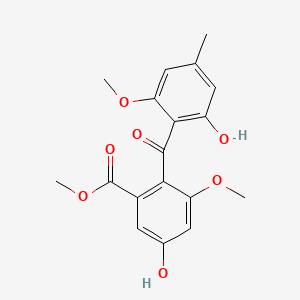 |
0.696 | D0D4HN | 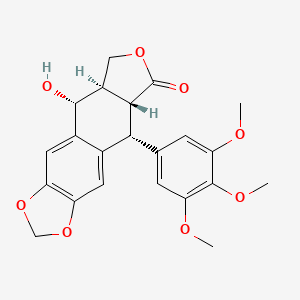 |
0.267 | ||
| ENC000936 | 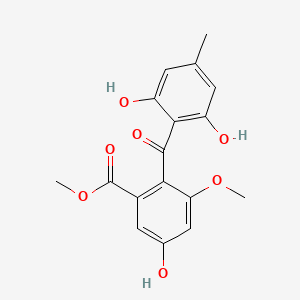 |
0.679 | D0W7JZ | 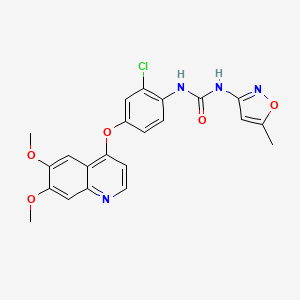 |
0.264 | ||
| ENC002683 | 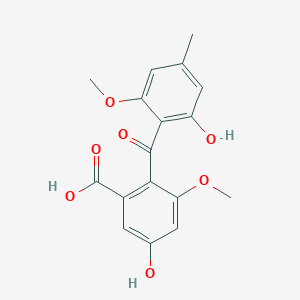 |
0.679 | D0AO5H | 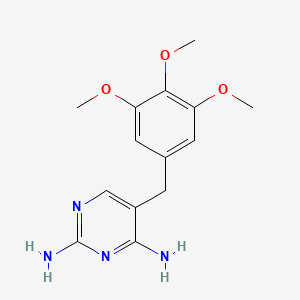 |
0.263 | ||
| ENC006012 | 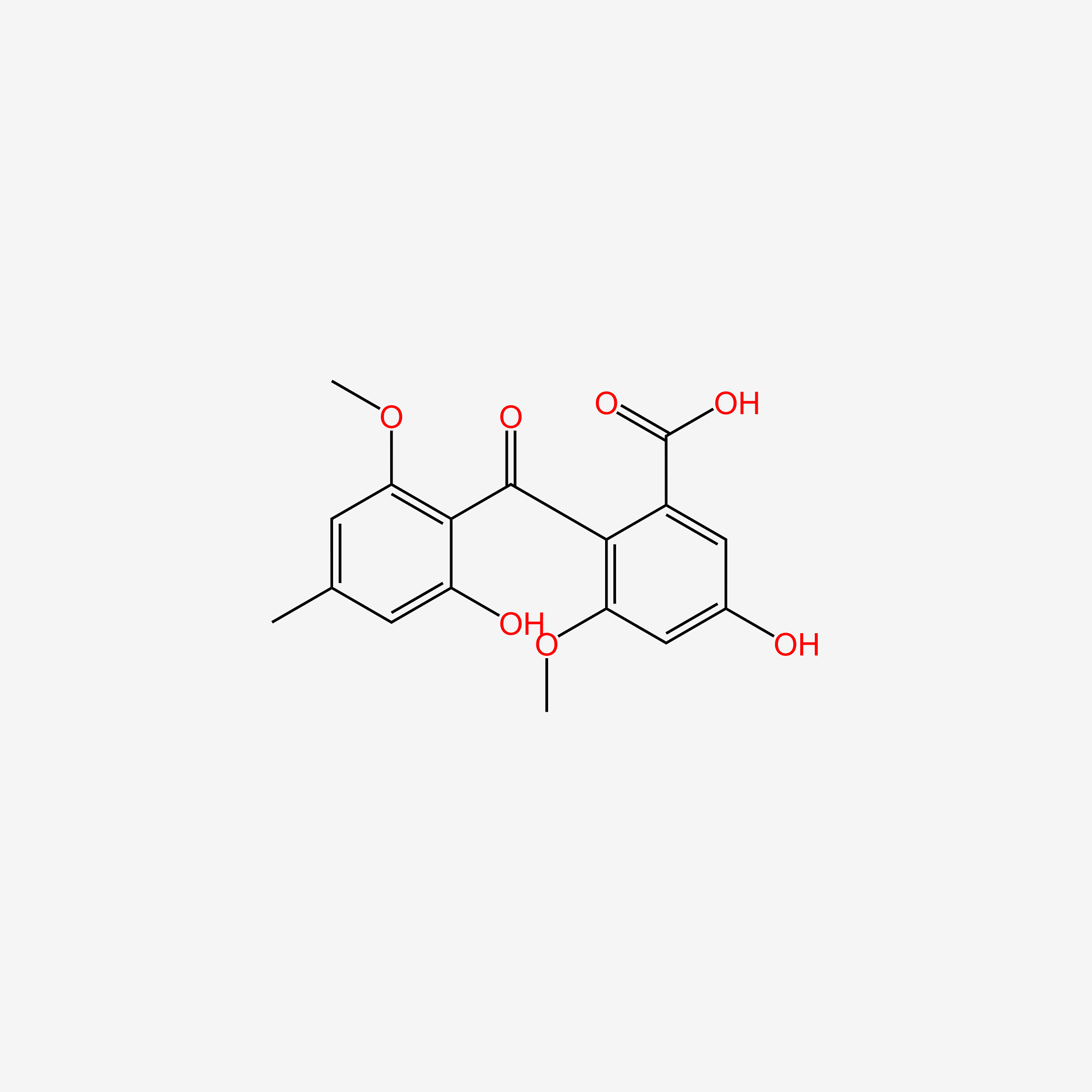 |
0.679 | D0N1FS | 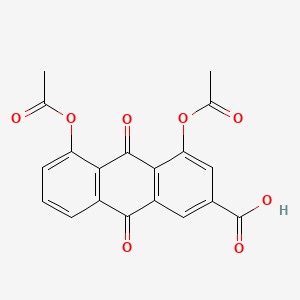 |
0.261 | ||