NPs Basic Information
|
Name |
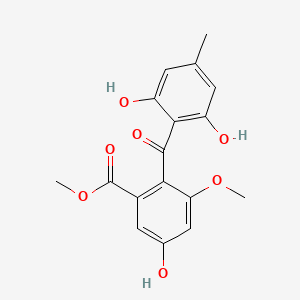
Sulochrin
|
| Molecular Formula | C17H16O7 | |
| IUPAC Name* |
methyl 2-(2,6-dihydroxy-4-methylbenzoyl)-5-hydroxy-3-methoxybenzoate
|
|
| SMILES |
CC1=CC(=C(C(=C1)O)C(=O)C2=C(C=C(C=C2OC)O)C(=O)OC)O
|
|
| InChI |
InChI=1S/C17H16O7/c1-8-4-11(19)15(12(20)5-8)16(21)14-10(17(22)24-3)6-9(18)7-13(14)23-2/h4-7,18-20H,1-3H3
|
|
| InChIKey |
YJRLSCDUYLRBIZ-UHFFFAOYSA-N
|
|
| Synonyms |
Sulochrin; 519-57-3; Methyl 2-(2,6-dihydroxy-4-methylbenzoyl)-5-hydroxy-3-methoxybenzoate; CHEBI:16159; 8NA53C271Z; Benzoic acid, 2-(2,6-dihydroxy-4-methylbenzoyl)-5-hydroxy-3-methoxy-, methyl ester; m-Anisic acid, 5-hydroxy-2-(4-methyl-.gamma.-resorcyloyl)-, methyl ester; m-Anisic acid, 5-hydroxy-2-(4-methyl-gamma-resorcyloyl)-, methyl ester; UNII-8NA53C271Z; Sulochrin_120096; MLS000863634; CHEMBL61133; SCHEMBL233392; MEGxm0_000192; ACon0_000926; ACon1_000208; DTXSID80199860; HMS2268H19; ZINC895263; Sulochrin, >=98% (HPLC), solid; NCGC00180780-01; SMR000440729; HY-105713; CS-0026511; C00495; BRD-K94458351-001-01-5; Q27098405; methyl 2-(2,6-dihydroxy-4-methyl-benzoyl)-5-hydroxy-3-methoxy-benzoate; Methyl 2-(2,6-dihydroxy-4-methylbenzoyl)-5-hydroxy-3-methoxybenzoate #
|
|
| CAS | 519-57-3 | |
| PubChem CID | 160505 | |
| ChEMBL ID | CHEMBL61133 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 332.3 | ALogp: | 2.9 |
| HBD: | 3 | HBA: | 7 |
| Rotatable Bonds: | 5 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 113.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 24 | QED Weighted: | 0.583 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.007 | MDCK Permeability: | 0.00001110 |
| Pgp-inhibitor: | 0.009 | Pgp-substrate: | 0.013 |
| Human Intestinal Absorption (HIA): | 0.093 | 20% Bioavailability (F20%): | 0.093 |
| 30% Bioavailability (F30%): | 0.78 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.049 | Plasma Protein Binding (PPB): | 97.63% |
| Volume Distribution (VD): | 0.504 | Fu: | 4.13% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.956 | CYP1A2-substrate: | 0.883 |
| CYP2C19-inhibitor: | 0.324 | CYP2C19-substrate: | 0.062 |
| CYP2C9-inhibitor: | 0.68 | CYP2C9-substrate: | 0.853 |
| CYP2D6-inhibitor: | 0.561 | CYP2D6-substrate: | 0.354 |
| CYP3A4-inhibitor: | 0.705 | CYP3A4-substrate: | 0.149 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 13.727 | Half-life (T1/2): | 0.754 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.031 | Human Hepatotoxicity (H-HT): | 0.045 |
| Drug-inuced Liver Injury (DILI): | 0.94 | AMES Toxicity: | 0.656 |
| Rat Oral Acute Toxicity: | 0.028 | Maximum Recommended Daily Dose: | 0.852 |
| Skin Sensitization: | 0.197 | Carcinogencity: | 0.022 |
| Eye Corrosion: | 0.004 | Eye Irritation: | 0.936 |
| Respiratory Toxicity: | 0.119 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC002468 | 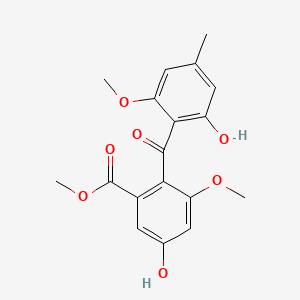 |
0.795 | D07MGA |  |
0.344 | ||
| ENC005978 | 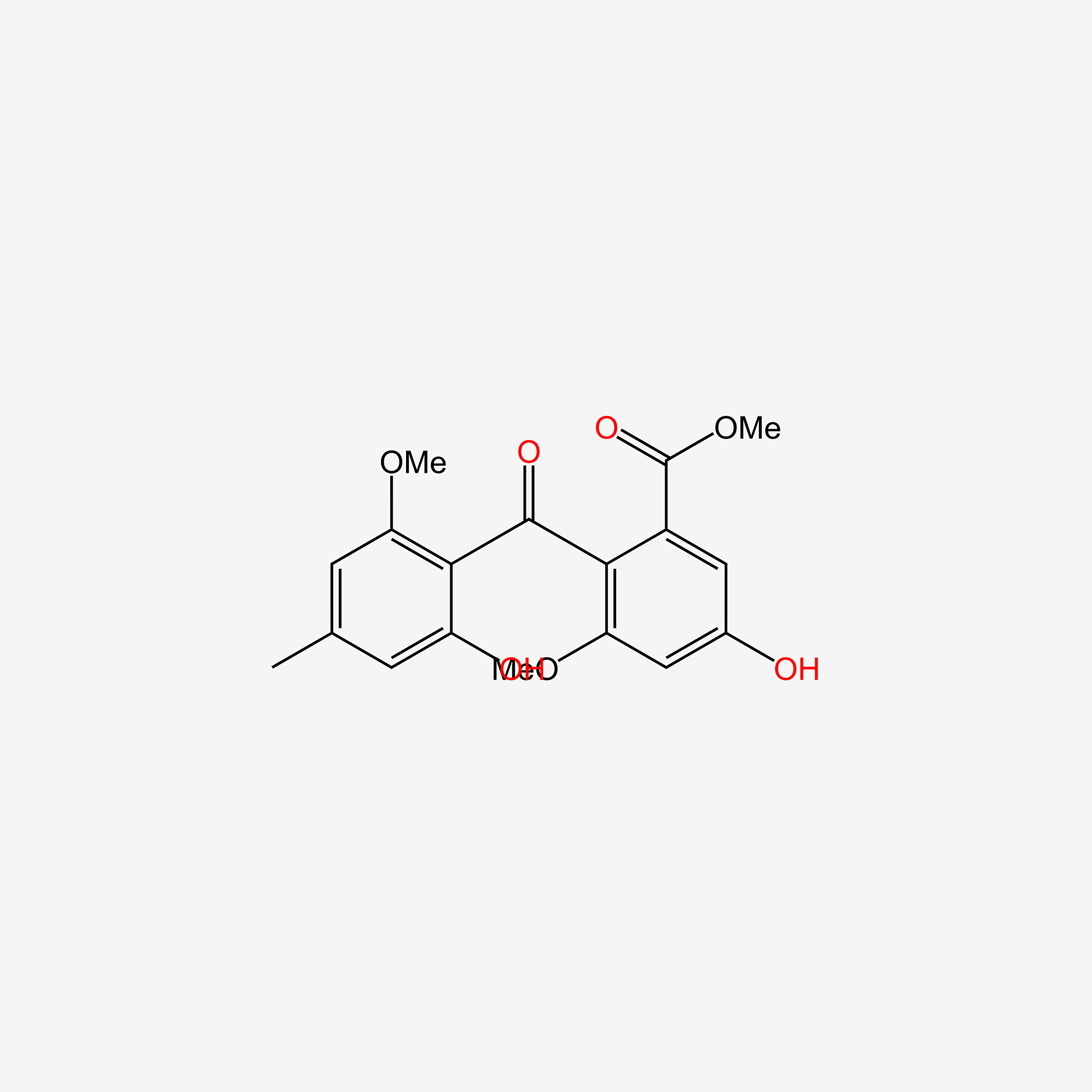 |
0.795 | D06GCK |  |
0.304 | ||
| ENC005979 | 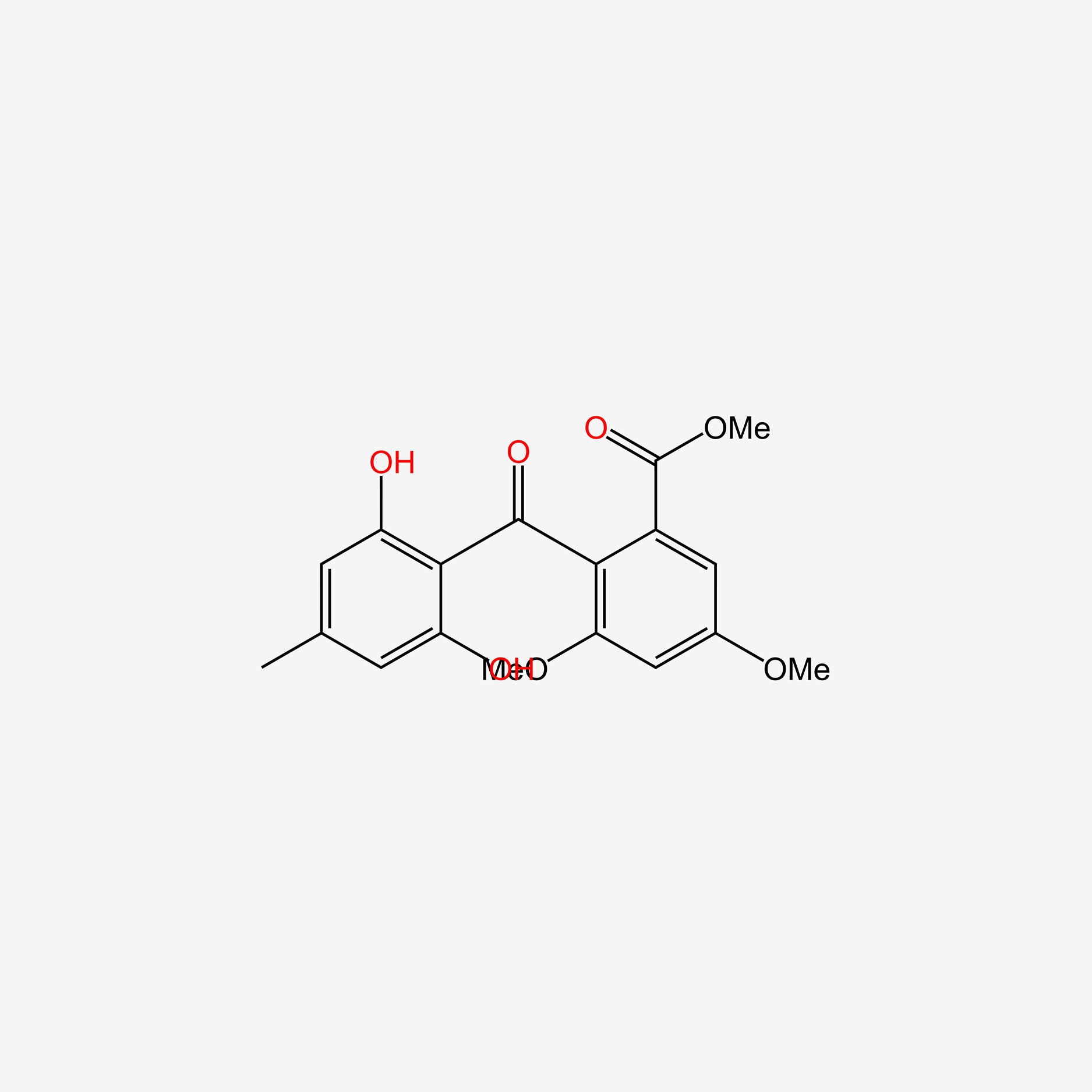 |
0.795 | D04AIT |  |
0.284 | ||
| ENC002375 | 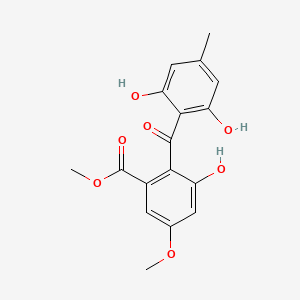 |
0.778 | D0Y7PG | 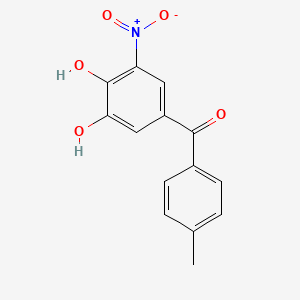 |
0.269 | ||
| ENC002109 | 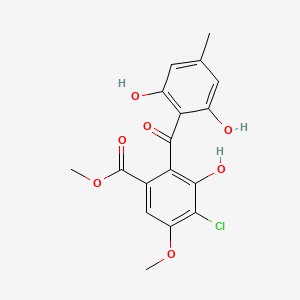 |
0.757 | D0K8KX | 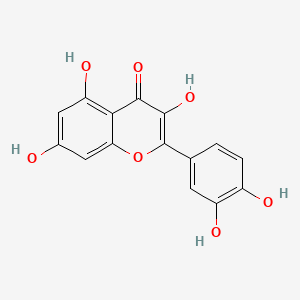 |
0.265 | ||
| ENC006012 | 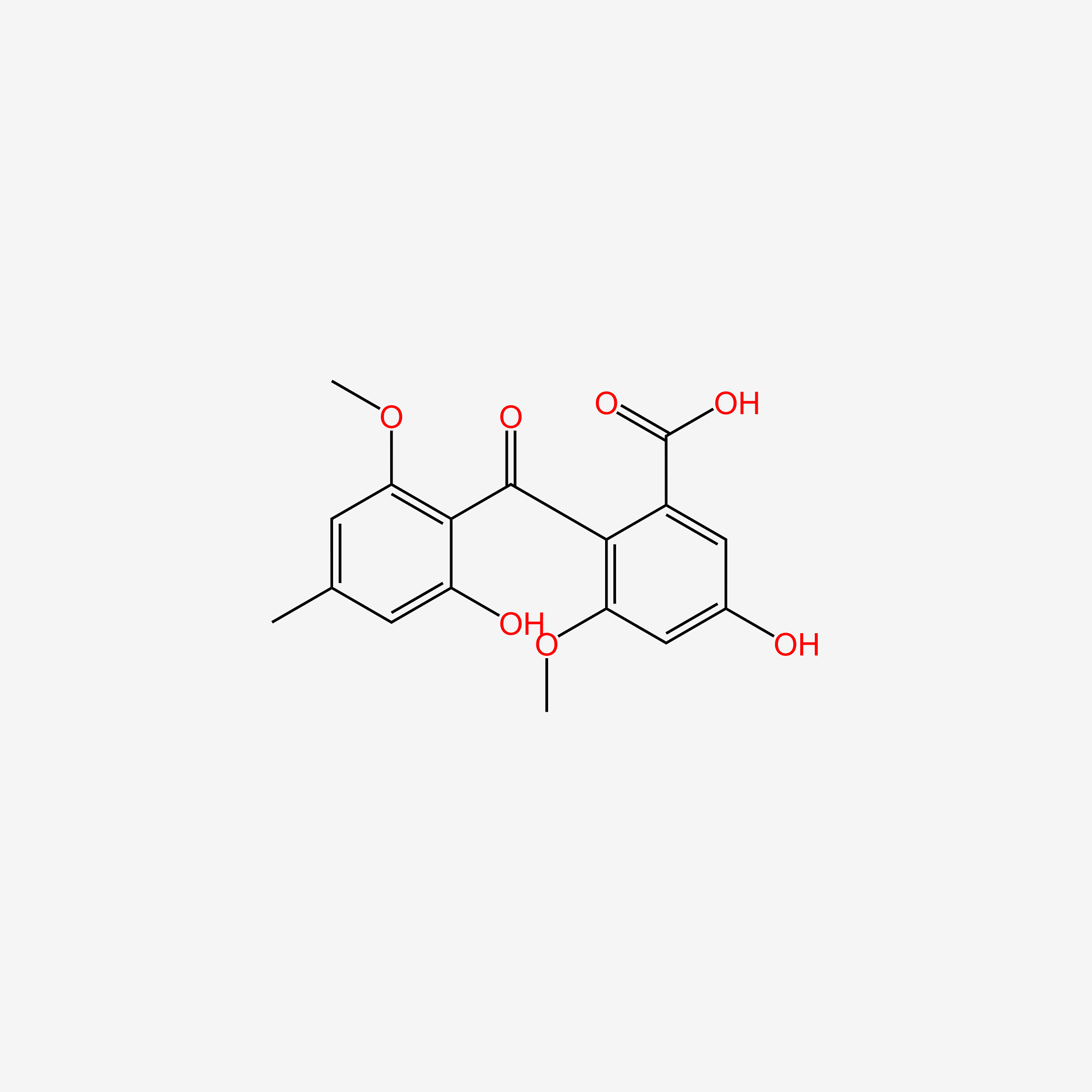 |
0.730 | D00KRE | 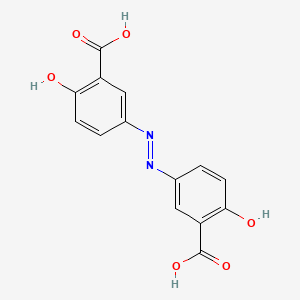 |
0.265 | ||
| ENC002683 | 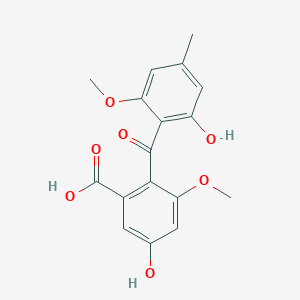 |
0.730 | D00WVW | 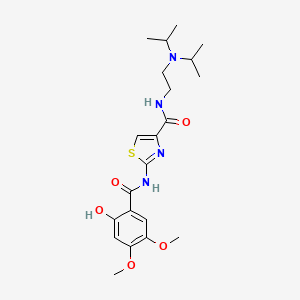 |
0.265 | ||
| ENC004806 | 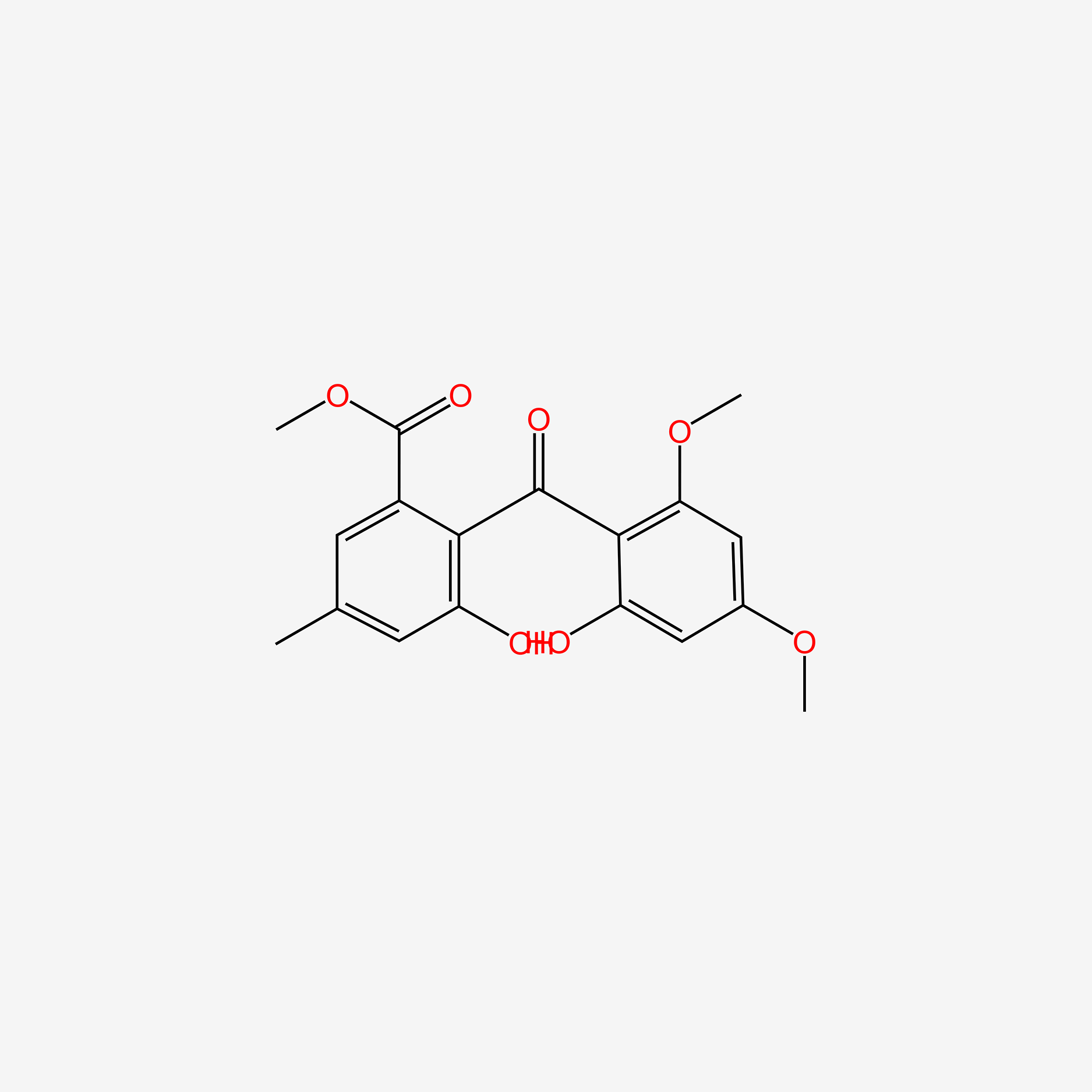 |
0.724 | D04OSE | 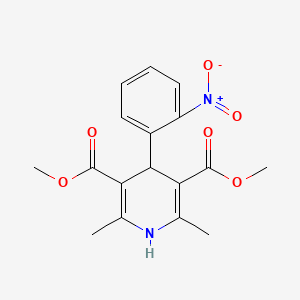 |
0.260 | ||
| ENC001490 | 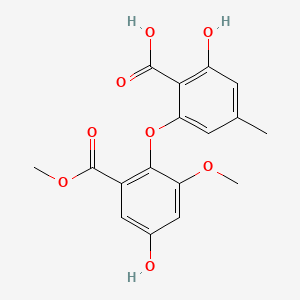 |
0.679 | D0E6OC | 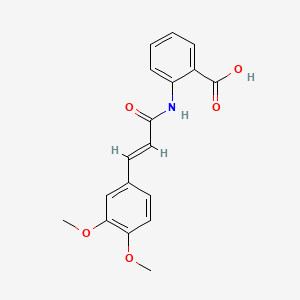 |
0.260 | ||
| ENC002461 | 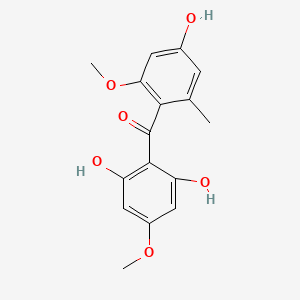 |
0.662 | D0AZ8C | 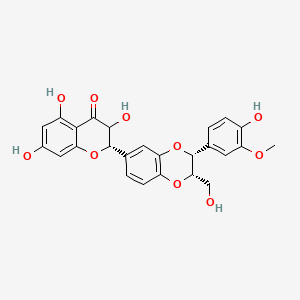 |
0.256 | ||