NPs Basic Information
|
Name |
Dihydrogeodin
|
| Molecular Formula | C17H14Cl2O7 | |
| IUPAC Name* |
methyl 2-(3,5-dichloro-2,6-dihydroxy-4-methylbenzoyl)-5-hydroxy-3-methoxybenzoate
|
|
| SMILES |
CC1=C(C(=C(C(=C1Cl)O)C(=O)C2=C(C=C(C=C2OC)O)C(=O)OC)O)Cl
|
|
| InChI |
InChI=1S/C17H14Cl2O7/c1-6-12(18)15(22)11(16(23)13(6)19)14(21)10-8(17(24)26-3)4-7(20)5-9(10)25-2/h4-5,20,22-23H,1-3H3
|
|
| InChIKey |
AXIPUIQLQUNOCF-UHFFFAOYSA-N
|
|
| Synonyms |
Dihydrogeodin; Geodin, dihydro-; 2151-16-8; Methyl 2-(3,5-dichloro-2,6-dihydroxy-4-methylbenzoyl)-5-hydroxy-3-methoxybenzoate; 2B275P7EST; m-Anisic acid, 2-(3,5-dichloro-4-methyl-.gamma.-resorcyloyl)-5-hydroxy-, methyl ester; Benzoic acid, 2-(3,5-dichloro-2,6-dihydroxy-4-methylbenzoyl)-5-hydroxy-3-methoxy-, methyl ester; 2-(2,6-Dihydroxy-3,5-dichloro-4-methylbenzoyl)-3-methoxy-5-hydroxybenzoic acid methyl ester; 2-[2,6-dihydroxy-3,5-dichloro-4-methylbenzoyl]-3-methoxy-5-hydroxybenzoic acid methyl ester; UNII-2B275P7EST; CHEMBL2035565; DTXSID10346502; CHEBI:155891; BS-1564; Q27896640; m-Anisic acid, 2-(3,5-dichloro-4-methyl-gamma-resorcyloyl)-5-hydroxy-, methyl ester; methyl 2-(3,5-dichloro-2,6-dihydroxy-4-methyl-benzoyl)-5-hydroxy-3-methoxy-benzoate; Methyl 2-(3,5-dichloro-2,6-dihydroxy-4-methylbenzoyl)-5-hydroxy-3-methoxybenzoate #; 2-(3,5-dichloro-2,6-dihydroxy-4-methylbenzoyl)-5-hydroxy-3-methoxybenzoic acid methyl ester
|
|
| CAS | 2151-16-8 | |
| PubChem CID | 612831 | |
| ChEMBL ID | CHEMBL2035565 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 401.2 | ALogp: | 4.2 |
| HBD: | 3 | HBA: | 7 |
| Rotatable Bonds: | 5 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 113.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 26 | QED Weighted: | 0.52 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.981 | MDCK Permeability: | 0.00001480 |
| Pgp-inhibitor: | 0.057 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.081 | 20% Bioavailability (F20%): | 0.007 |
| 30% Bioavailability (F30%): | 0.008 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.01 | Plasma Protein Binding (PPB): | 100.85% |
| Volume Distribution (VD): | 0.278 | Fu: | 2.03% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.878 | CYP1A2-substrate: | 0.938 |
| CYP2C19-inhibitor: | 0.28 | CYP2C19-substrate: | 0.078 |
| CYP2C9-inhibitor: | 0.848 | CYP2C9-substrate: | 0.816 |
| CYP2D6-inhibitor: | 0.382 | CYP2D6-substrate: | 0.248 |
| CYP3A4-inhibitor: | 0.298 | CYP3A4-substrate: | 0.142 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.257 | Half-life (T1/2): | 0.357 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.02 | Human Hepatotoxicity (H-HT): | 0.109 |
| Drug-inuced Liver Injury (DILI): | 0.962 | AMES Toxicity: | 0.069 |
| Rat Oral Acute Toxicity: | 0.035 | Maximum Recommended Daily Dose: | 0.916 |
| Skin Sensitization: | 0.377 | Carcinogencity: | 0.039 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.919 |
| Respiratory Toxicity: | 0.197 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
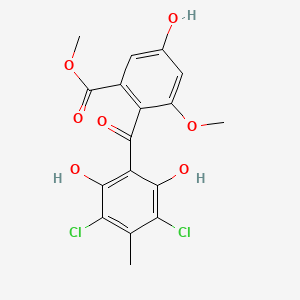
| ENC001415 | 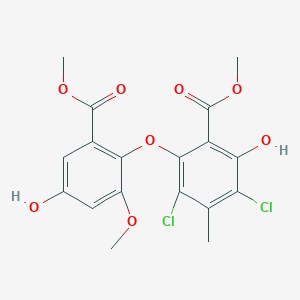 |
0.651 | D0WN0U | 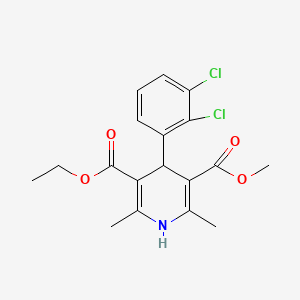 |
0.286 | ||
| ENC000936 | 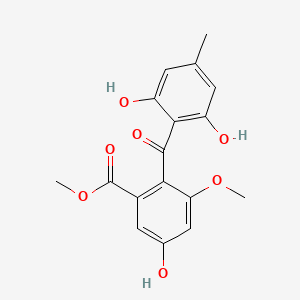 |
0.630 | D06GCK |  |
0.280 | ||
| ENC002109 | 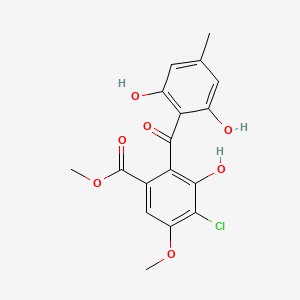 |
0.614 | D0WY9N | 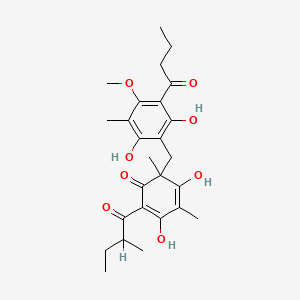 |
0.268 | ||
| ENC005978 | 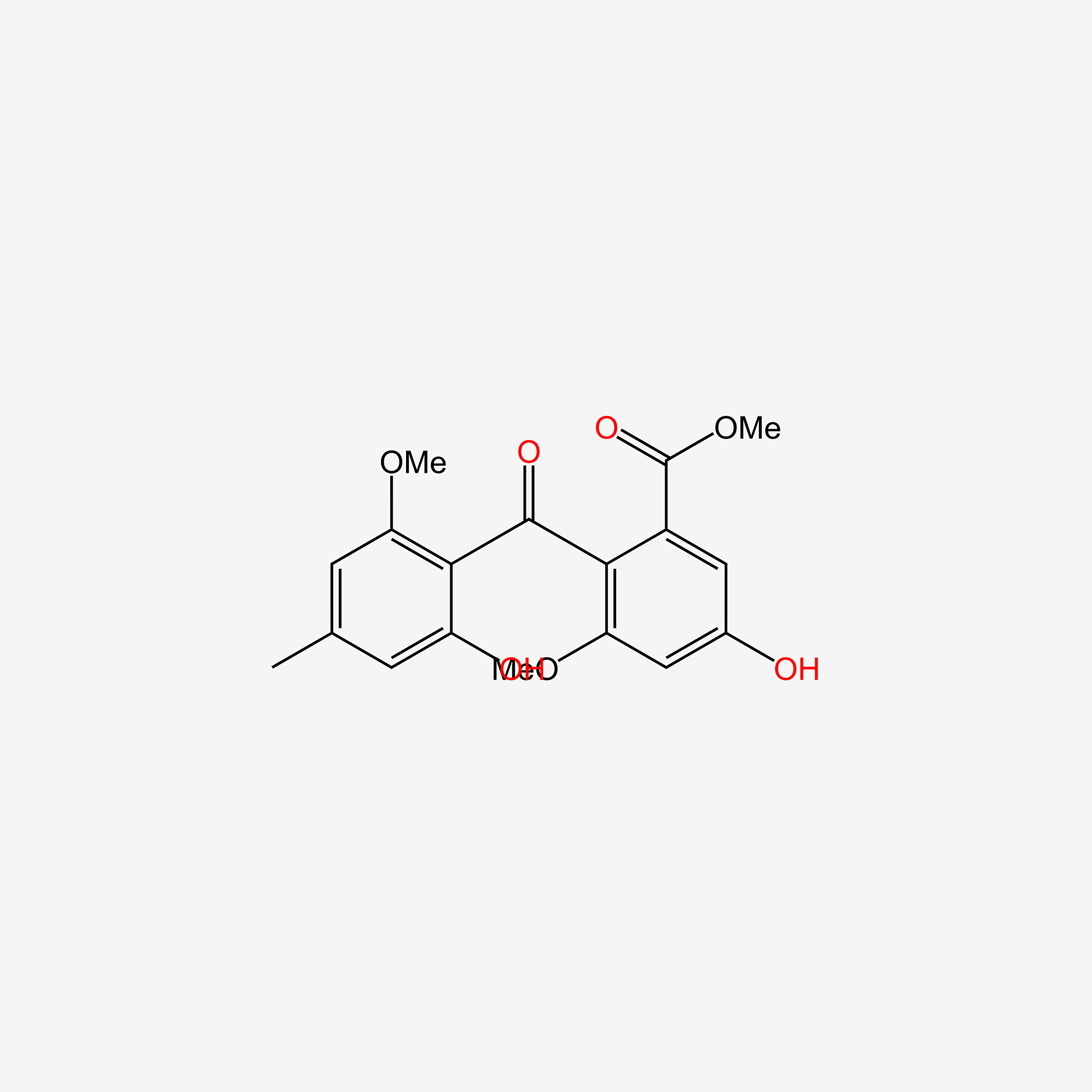 |
0.570 | D0ZX2G | 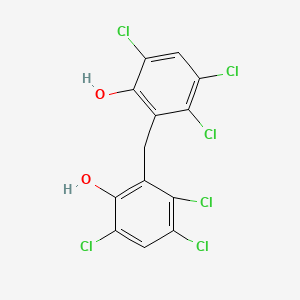 |
0.268 | ||
| ENC002468 | 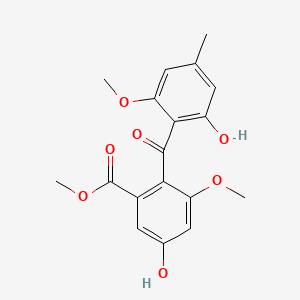 |
0.570 | D07MGA |  |
0.265 | ||
| ENC004226 | 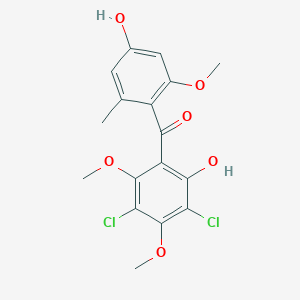 |
0.558 | D09ELP | 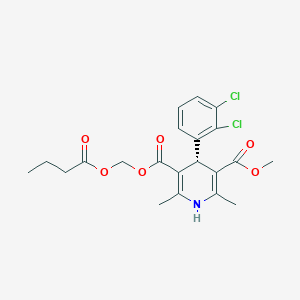 |
0.252 | ||
| ENC002470 | 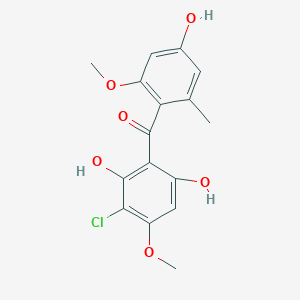 |
0.554 | D0QD1G | 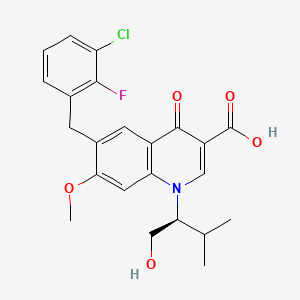 |
0.246 | ||
| ENC002683 | 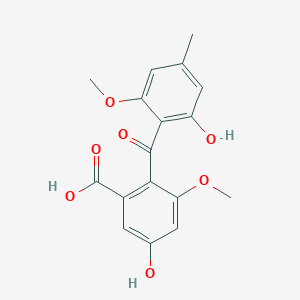 |
0.517 | D0C1SF | 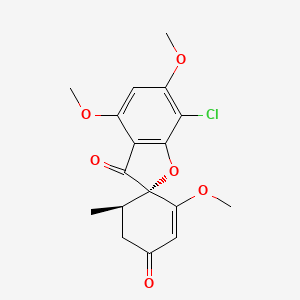 |
0.243 | ||
| ENC006012 | 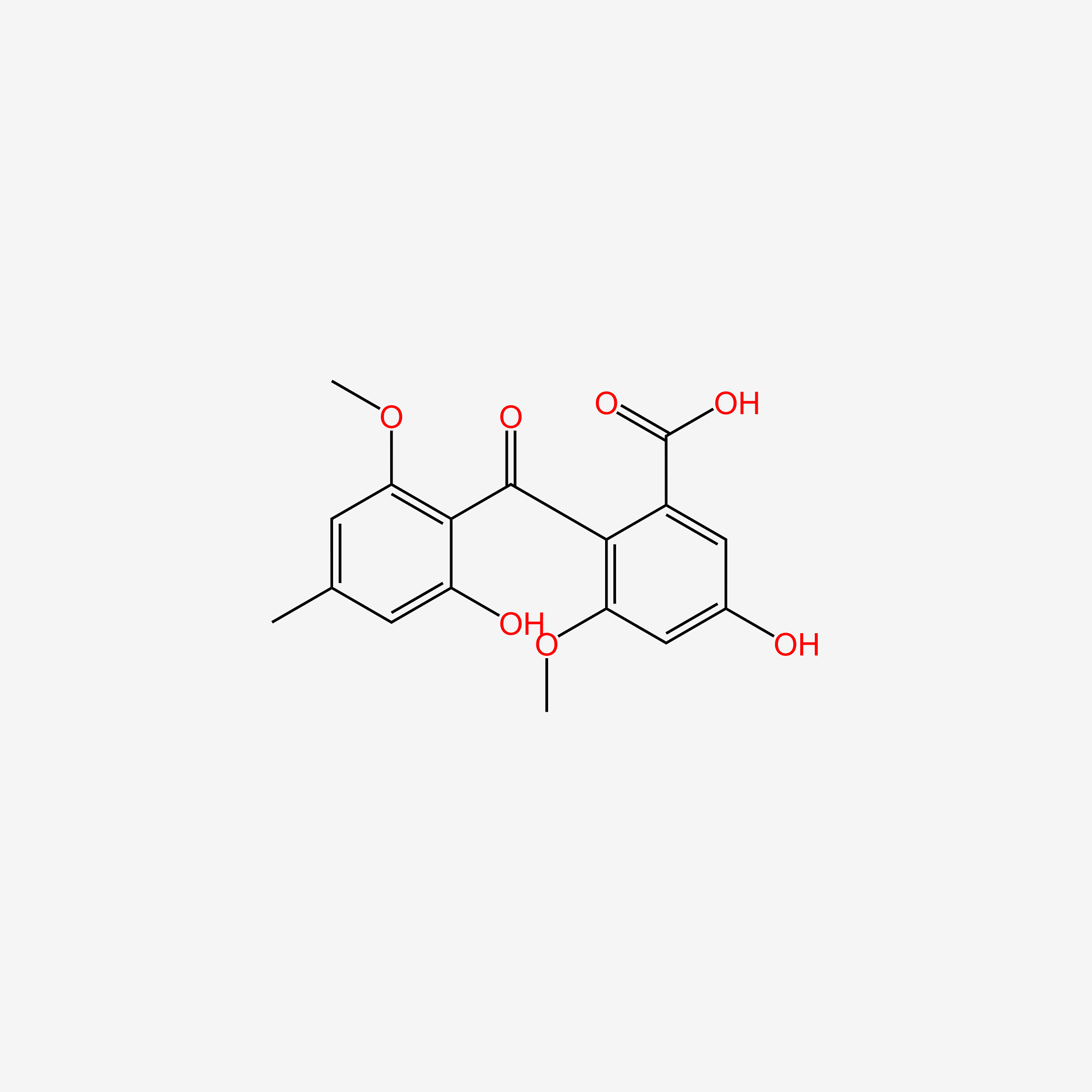 |
0.517 | D06TNL | 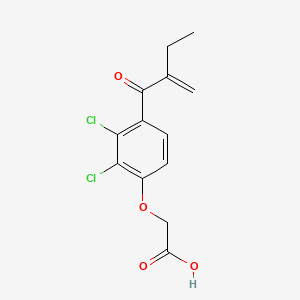 |
0.242 | ||
| ENC005979 | 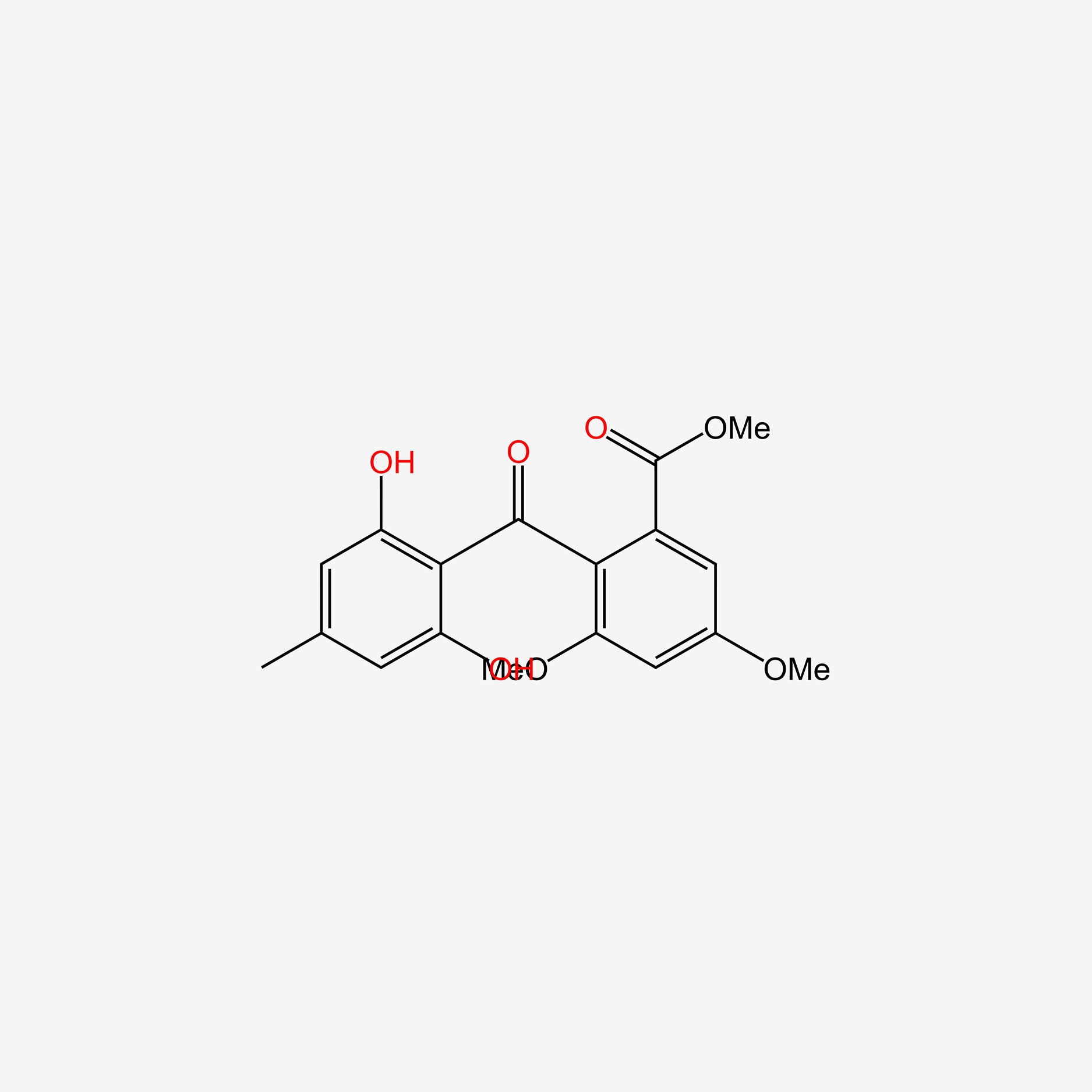 |
0.500 | D00WVW | 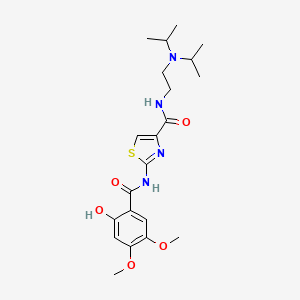 |
0.236 | ||