NPs Basic Information
|
Name |
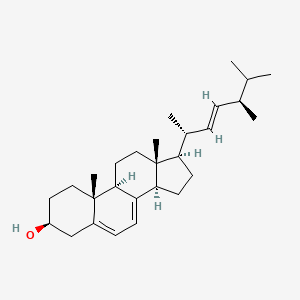
Ergosterol
|
| Molecular Formula | C28H44O | |
| IUPAC Name* |
(3S,9S,10R,13R,14R,17R)-17-[(E,2R,5R)-5,6-dimethylhept-3-en-2-yl]-10,13-dimethyl-2,3,4,9,11,12,14,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-ol
|
|
| SMILES |
C[C@H](/C=C/[C@H](C)C(C)C)[C@H]1CC[C@@H]2[C@@]1(CC[C@H]3C2=CC=C4[C@@]3(CC[C@@H](C4)O)C)C
|
|
| InChI |
InChI=1S/C28H44O/c1-18(2)19(3)7-8-20(4)24-11-12-25-23-10-9-21-17-22(29)13-15-27(21,5)26(23)14-16-28(24,25)6/h7-10,18-20,22,24-26,29H,11-17H2,1-6H3/b8-7+/t19-,20+,22-,24+,25-,26-,27-,28+/m0/s1
|
|
| InChIKey |
DNVPQKQSNYMLRS-APGDWVJJSA-N
|
|
| Synonyms |
ERGOSTEROL; 57-87-4; Provitamin D2; Ergosterin; Ergosta-5,7,22-trien-3-ol, (3b,22E)-; Ergosta-5,7,22E-trien-3beta-ol; Z30RAY509F; CHEBI:16933; (22E,24S)-24-methylcholesta-5,7,22-trien-3beta-ol; (22E)-ergosta-5,7,22-trien-3beta-ol; (22E)-Ergosta-5,7,22-trien-3.beta.-ol; 24-Methylcholesta-5,7,22-trien-3beta-ol; Ergosta-5,7,22-trien-3beta-ol; (3S,9S,10R,13R,14R,17R)-17-[(E,2R,5R)-5,6-dimethylhept-3-en-2-yl]-10,13-dimethyl-2,3,4,9,11,12,14,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-ol; (3.beta.,22E)-Ergosta-5,7,22-trien-3-ol; Ergosta-5,7,22-trien-3-ol, (3.beta.,22E)-; ergosta-5,7,22-trien-3-ol; UNII-Z30RAY509F; NSC62791; CCRIS 7220; HSDB 395; Provitamine D2; 5,7,22-Ergostatrien-3beta-ol; Ergosta-5,7,22-trien-3-ol, (3beta,22E)-; (3S,9S,10R,13R,14R,17R)-10,13-dimethyl-17-[(E,1R,4R)-1,4,5-trimethylhex-2-enyl]-2,3,4,9,11,12,14,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-ol; EINECS 200-352-7; 3beta-Hydroxy-5,7,22-ergostatriene; 24alpha-Methyl-22E-dehydrocholesterol; Ergosterol, >=75%; ERGOSTEROL [MI]; AI3-18876; ERGOSTEROL [HSDB]; ERGOSTEROL [INCI]; 24R-Methylcholesta-5,7,2E-trien-3beta-ol; bmse000494; Ergosterol (Provitamin D2); ERGOSTEROL [USP-RS]; SCHEMBL43194; MEGxm0_000450; CHEMBL1232562; ACon0_000429; ACon1_000637; DTXSID90878679; 24a-Methyl-22E-dehydrocholesterol; Ergosterol, >=95.0% (HPLC); HY-N0181; ZINC4084618; BDBM50378884; LMST01030093; s2297; 24alpha-Methyl-22E-dehydrocholestero; AKOS015918128; AC-8370; DB04038; DS-4956; (24R)-Ergosta-5,7,22-trien-3b-ol; (3beta)-Ergosta-5,7,22-trien-3-ol; NCGC00168889-01; NCGC00168889-02; (3S,9S,10R,13R,14R,17R)-17-((2R,5R,E)-5,6-dimethylhept-3-en-2-yl)-10,13-dimethyl-2,3,4,9,10,11,12,13,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol; 14-((2E)(1R,4R)-1,4,5-trimethylhex-2-enyl)(1S,5S,2R,11R,14R,15R)-2,15-dimethyl tetracyclo[8.7.0.0<2,7>.0<11,15>]heptadeca-7,9-dien-5-ol; 24-Methylcholesta-5,7,22-trien-3b-ol; 24R-Methylcholesta-5,7,22E-trien-3b-ol; (3beta,2E)-Ergosta-5,7,22-trien-3-ol; CS-0007890; (3beta,22E)-Ergosta-5,7,22-trien-3-ol; 24R-Methylcholesta-5,7,22E-trien-3beta-ol; C01694; ERGOCALCIFEROL IMPURITY B [EP IMPURITY]; Ergosta-5,7,22-trien-3-ol,(3|A,22E)-; 003E623; EN300-21680273; Q143263; 45ED0A4C-6FDA-443F-B886-D6C805A76AF2; Ergosterol, 10 mg/mL in chloroform, analytical standard; (3beta,14beta,17alpha,22E)-ergosta-5,7,22-trien-3-ol; Ergosterol, European Pharmacopoeia (EP) Reference Standard; Ergosterol, United States Pharmacopeia (USP) Reference Standard; ERGOSTEROL (CONSTITUENT OF GANODERMA LUCIDUM FRUITING BODY) [DSC]; Ergosterol, Pharmaceutical Secondary Standard; Certified Reference Material; (1R,3aR,7S,9aR,9bS,11aR)-1-[(2R,3E,5R)-5,6-dimethylhept-3-en-2-yl]-9a,11a-dimethyl-1H,2H,3H,3aH,6H,7H,8H,9H,9aH,9bH,10H,11H,11aH-cyclopenta[a]phenanthren-7-ol
|
|
| CAS | 57-87-4 | |
| PubChem CID | 444679 | |
| ChEMBL ID | CHEMBL1232562 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 396.6 | ALogp: | 7.4 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 4 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 20.2 | Aromatic Rings: | 4 |
| Heavy Atoms: | 29 | QED Weighted: | 0.491 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.815 | MDCK Permeability: | 0.00001130 |
| Pgp-inhibitor: | 0.996 | Pgp-substrate: | 0.381 |
| Human Intestinal Absorption (HIA): | 0.015 | 20% Bioavailability (F20%): | 0.949 |
| 30% Bioavailability (F30%): | 0.921 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.214 | Plasma Protein Binding (PPB): | 93.37% |
| Volume Distribution (VD): | 1.32 | Fu: | 1.79% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.08 | CYP1A2-substrate: | 0.425 |
| CYP2C19-inhibitor: | 0.08 | CYP2C19-substrate: | 0.961 |
| CYP2C9-inhibitor: | 0.212 | CYP2C9-substrate: | 0.052 |
| CYP2D6-inhibitor: | 0.022 | CYP2D6-substrate: | 0.137 |
| CYP3A4-inhibitor: | 0.431 | CYP3A4-substrate: | 0.921 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 1.065 | Half-life (T1/2): | 0.121 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.023 | Human Hepatotoxicity (H-HT): | 0.126 |
| Drug-inuced Liver Injury (DILI): | 0.021 | AMES Toxicity: | 0.005 |
| Rat Oral Acute Toxicity: | 0.96 | Maximum Recommended Daily Dose: | 0.982 |
| Skin Sensitization: | 0.851 | Carcinogencity: | 0.013 |
| Eye Corrosion: | 0.004 | Eye Irritation: | 0.185 |
| Respiratory Toxicity: | 0.97 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC004738 | 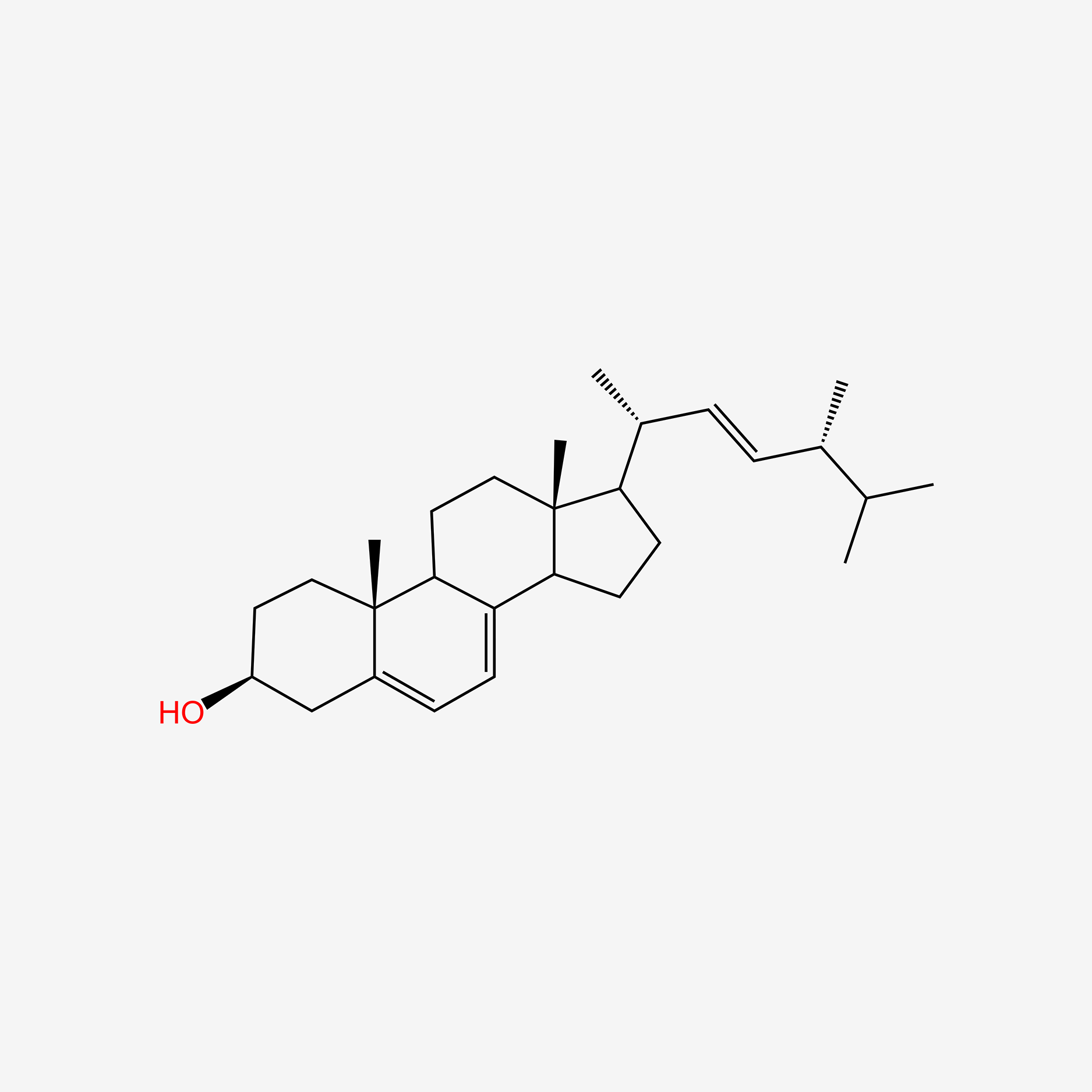 |
1.000 | D0G8OC | 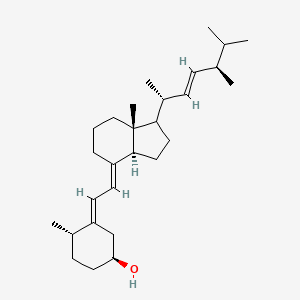 |
0.553 | ||
| ENC005707 | 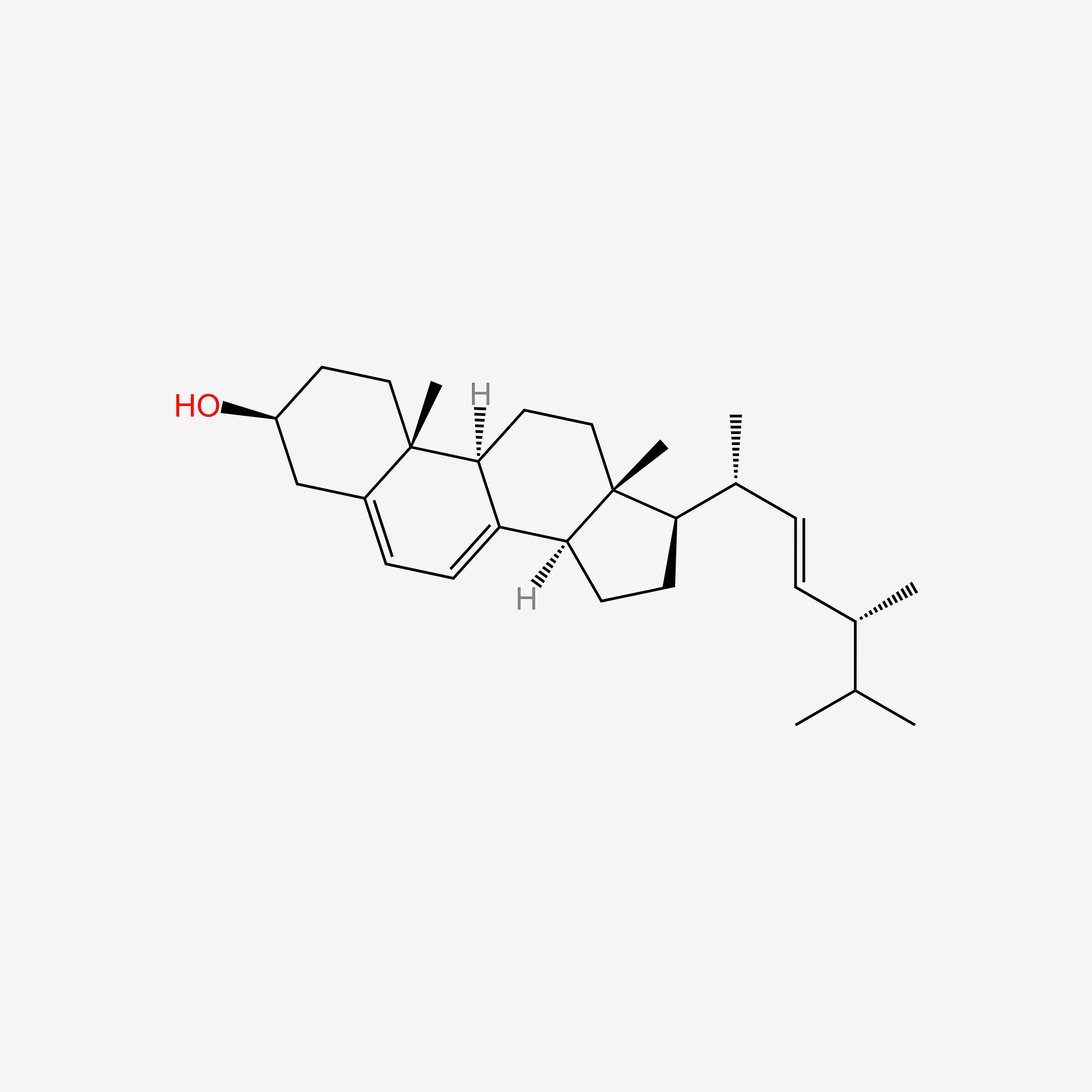 |
1.000 | D06JPB |  |
0.509 | ||
| ENC005258 | 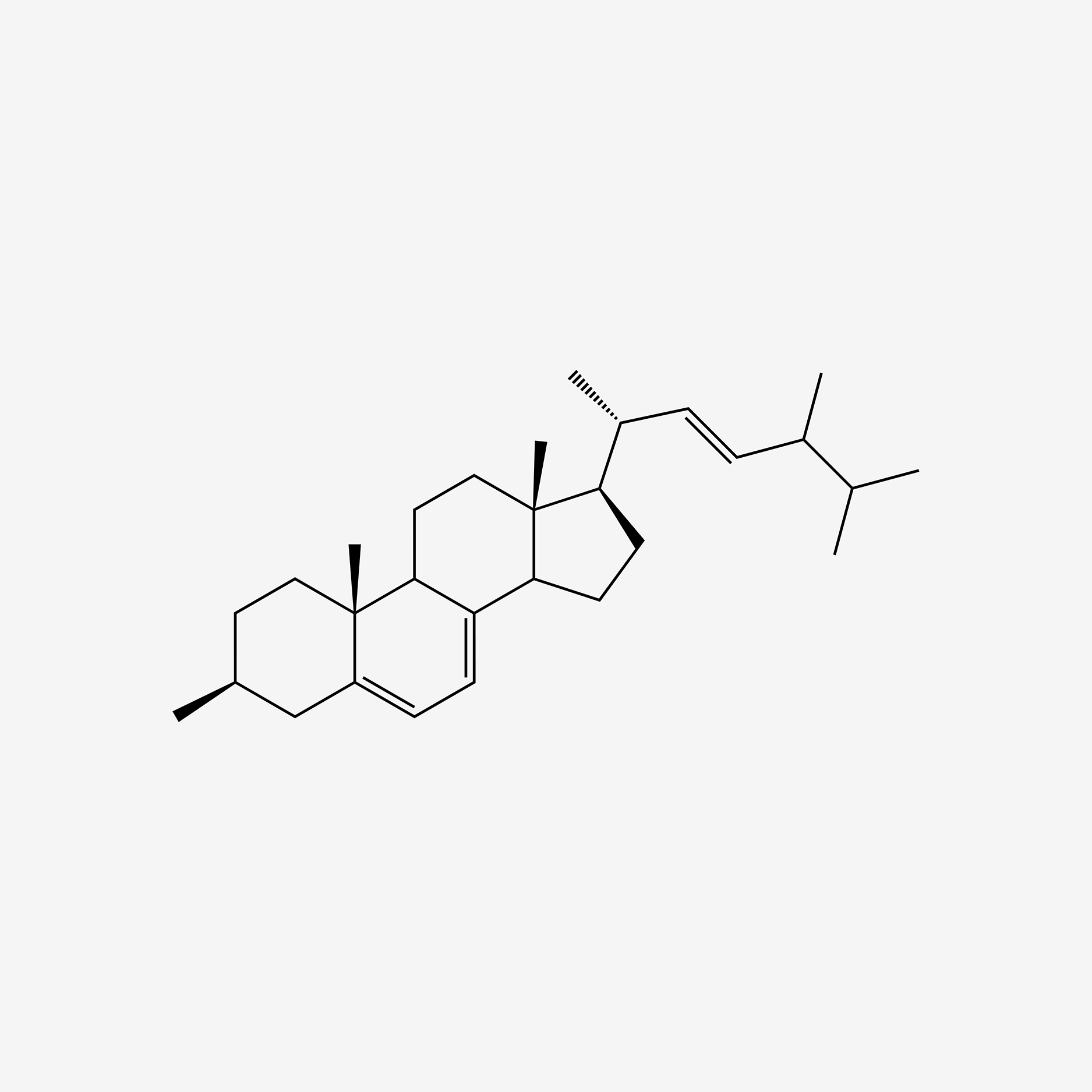 |
0.860 | D0G5CF | 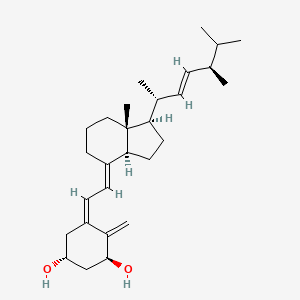 |
0.486 | ||
| ENC005238 | 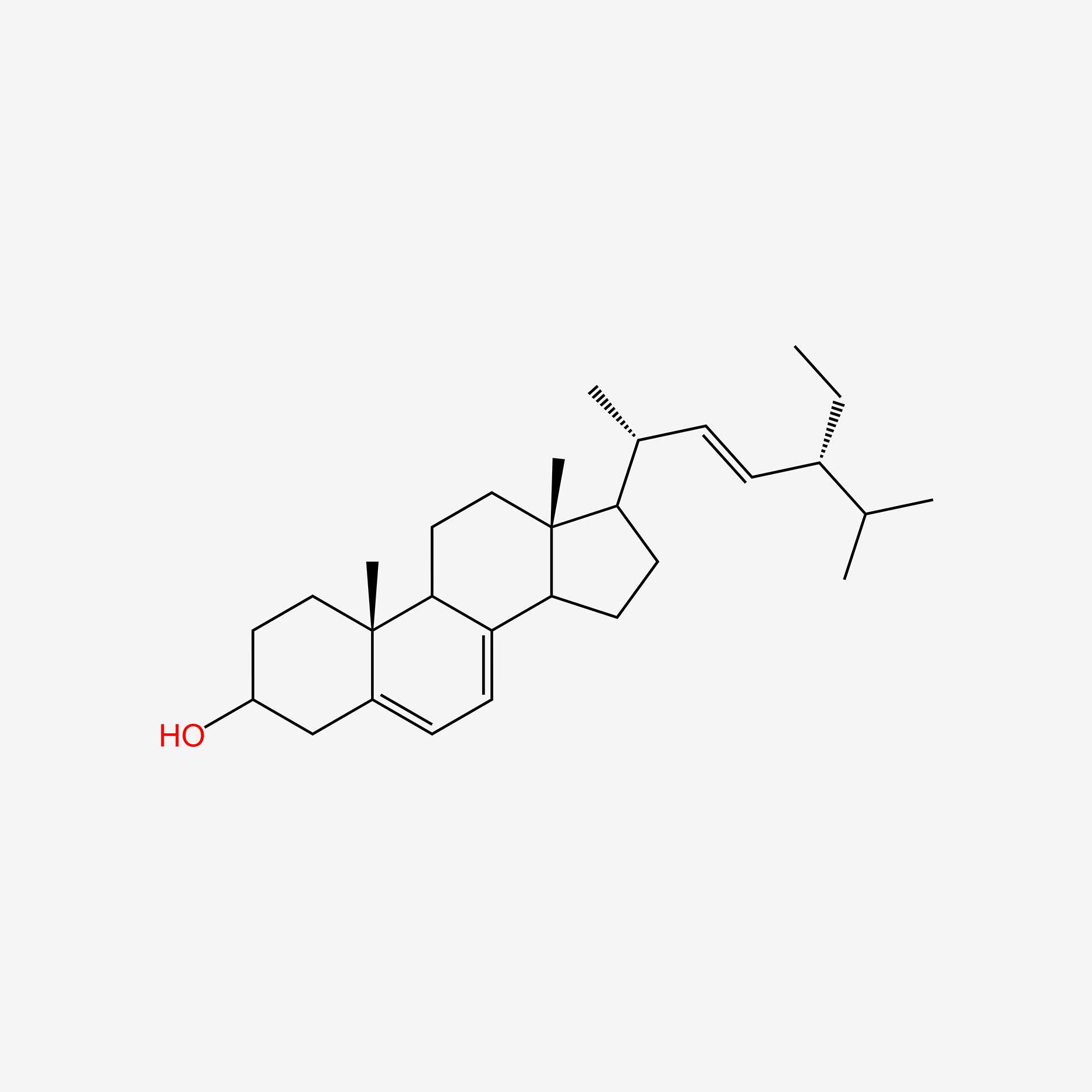 |
0.852 | D0Y7LD | 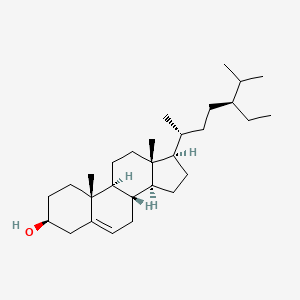 |
0.442 | ||
| ENC005270 | 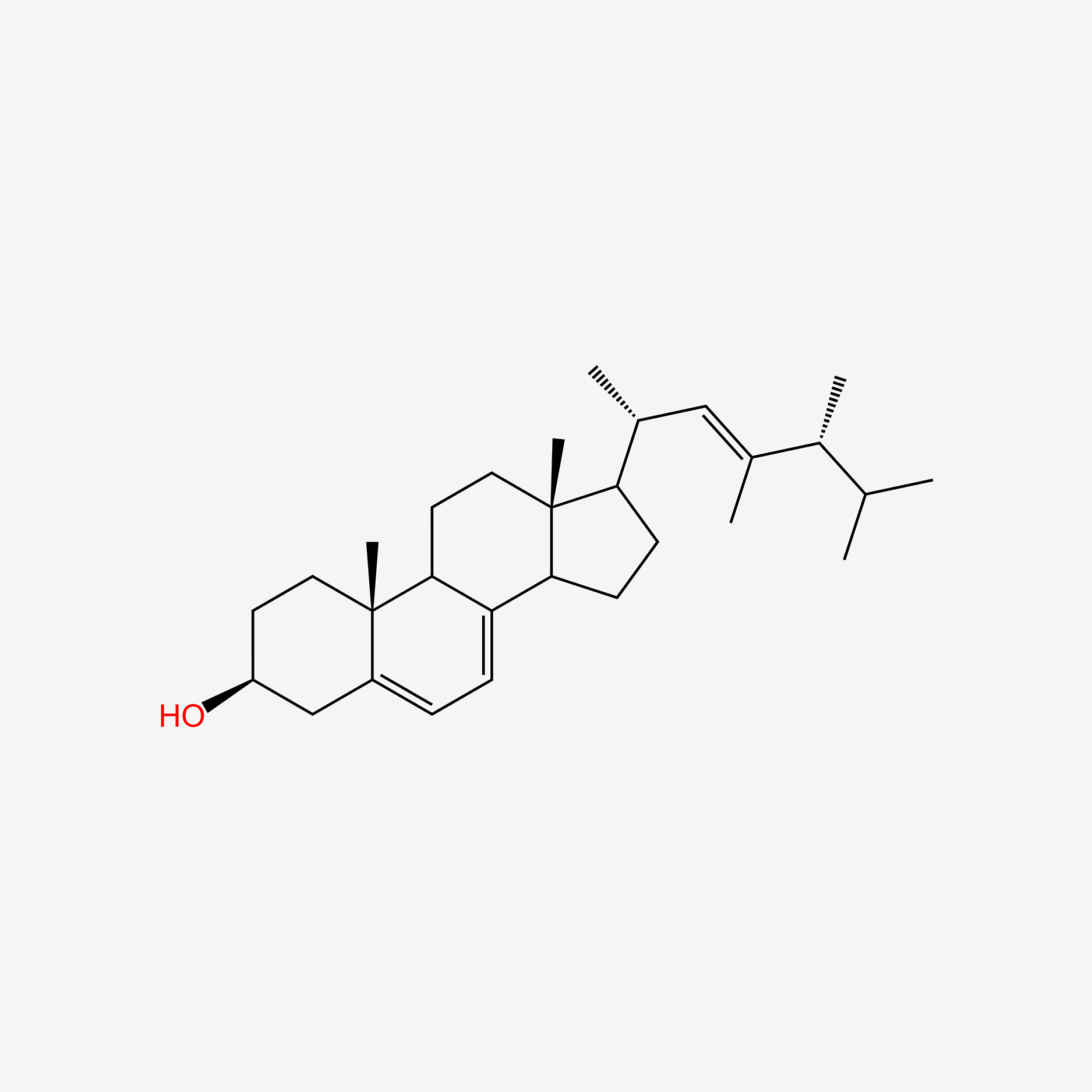 |
0.780 | D0N1TP | 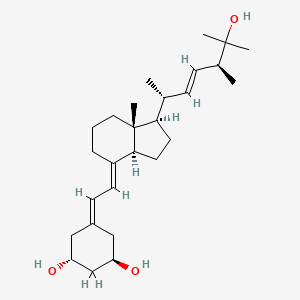 |
0.385 | ||
| ENC002892 | 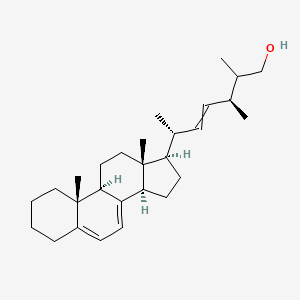 |
0.677 | D0B4RU | 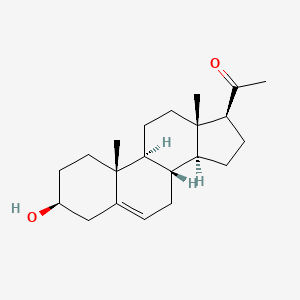 |
0.358 | ||
| ENC004735 | 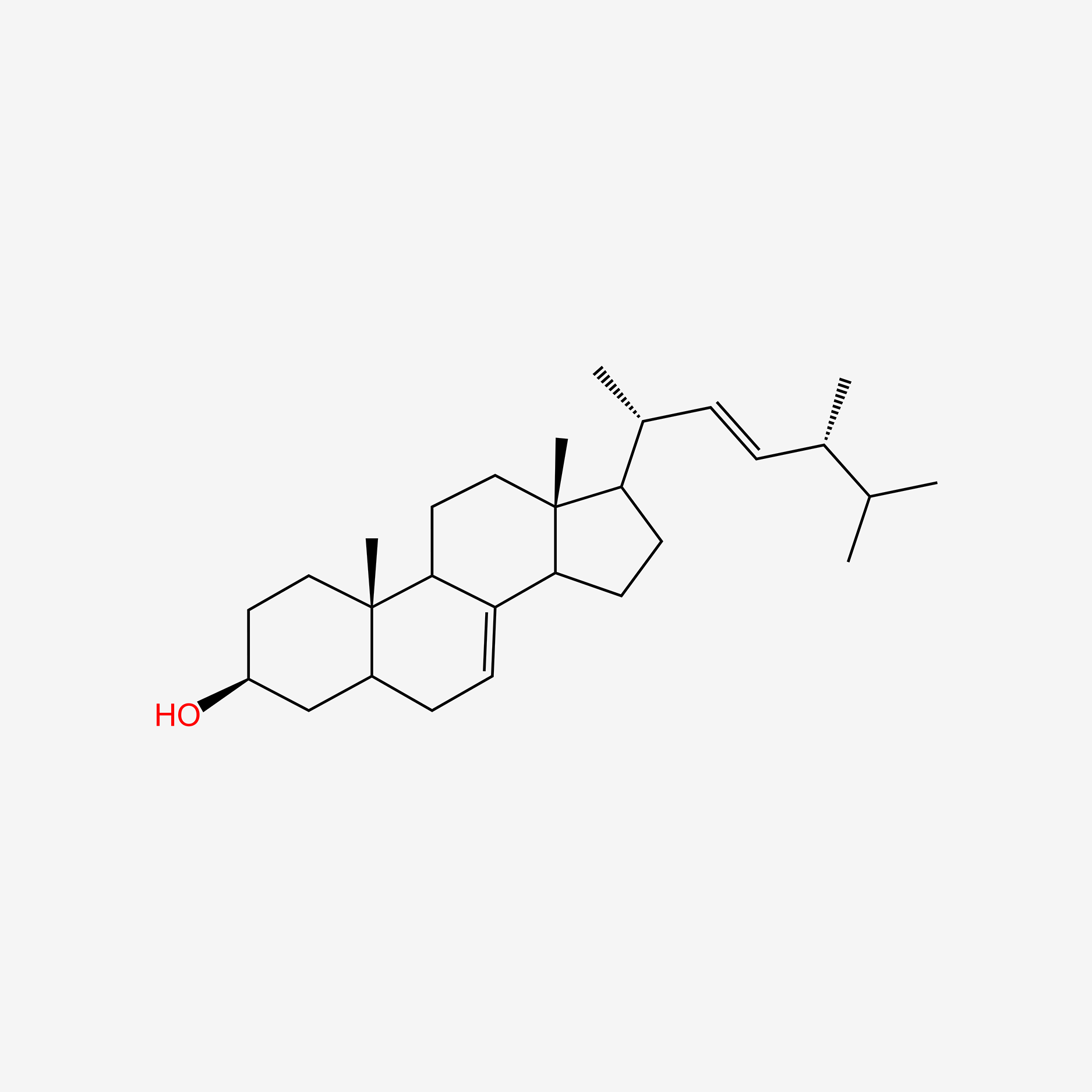 |
0.667 | D01QUS | 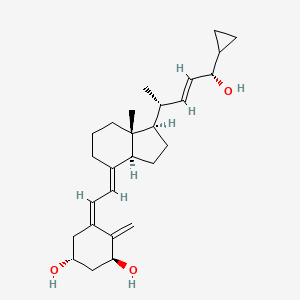 |
0.358 | ||
| ENC004803 | 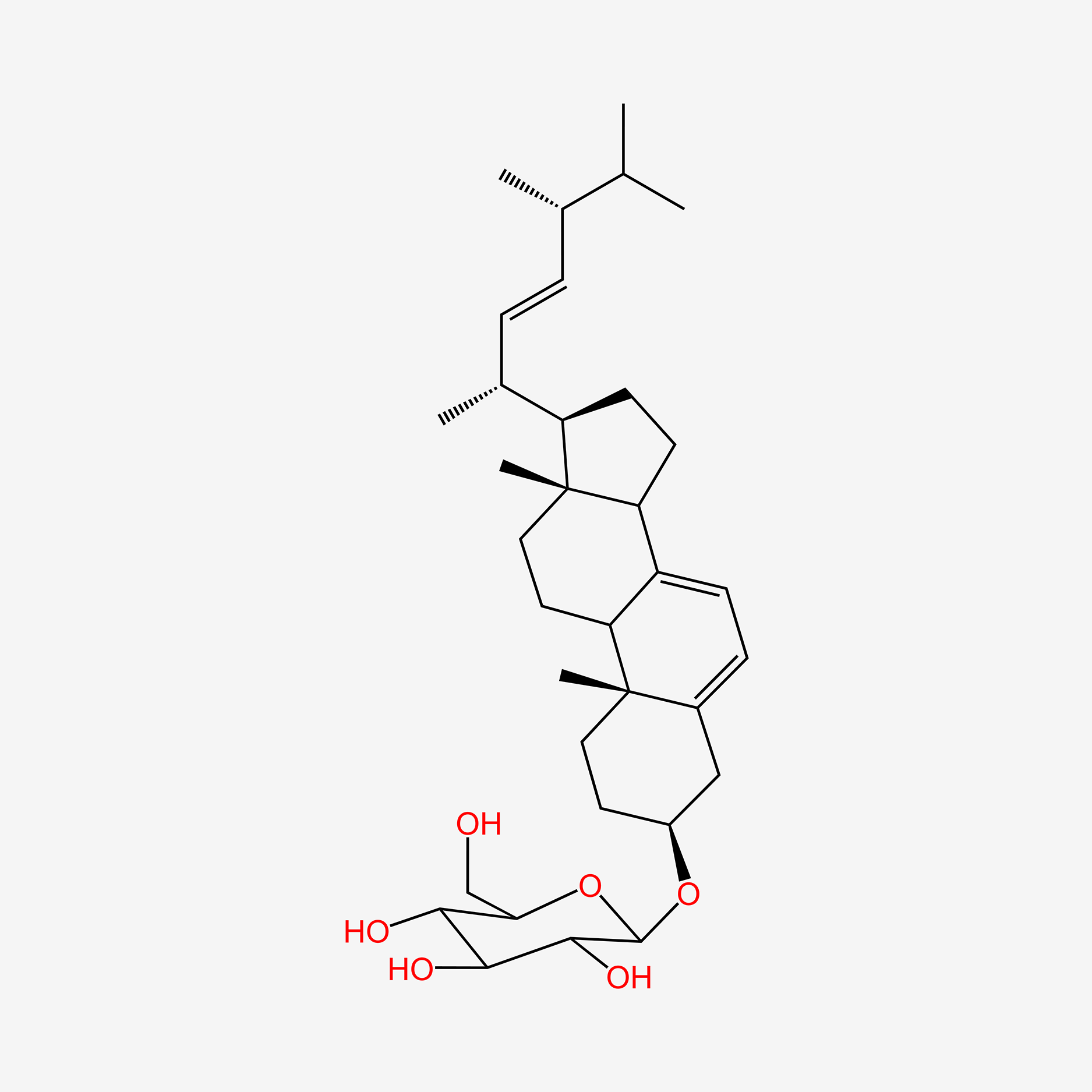 |
0.667 | D0K5WS |  |
0.339 | ||
| ENC002665 | 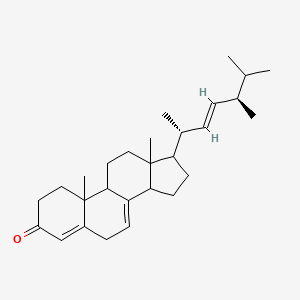 |
0.667 | D08SVH |  |
0.339 | ||
| ENC004758 |  |
0.649 | D0K0EK |  |
0.337 | ||