NPs Basic Information
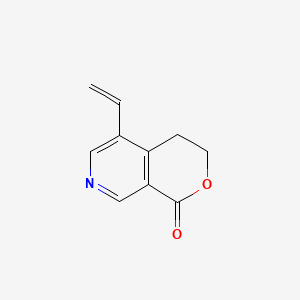
|
Name |
Gentianine
|
| Molecular Formula | C10H9NO2 | |
| IUPAC Name* |
5-ethenyl-3,4-dihydropyrano[3,4-c]pyridin-1-one
|
|
| SMILES |
C=CC1=CN=CC2=C1CCOC2=O
|
|
| InChI |
InChI=1S/C10H9NO2/c1-2-7-5-11-6-9-8(7)3-4-13-10(9)12/h2,5-6H,1,3-4H2
|
|
| InChIKey |
DFNZYFAJQPLJFI-UHFFFAOYSA-N
|
|
| Synonyms |
Gentianine; 439-89-4; Gentiannine; 5-ethenyl-3,4-dihydropyrano[3,4-c]pyridin-1-one; 5-ethenyl-3,4-dihydro-1H-pyrano[3,4-c]pyridin-1-one; C2PD310UXB; NSC606848; NSC-606848; 1H-Pyrano[3,4-c]pyridin-1-one, 5-ethenyl-3,4-dihydro-; 5-ethenyl-1H,3H,4H-pyrano[3,4-c]pyridin-1-one; 4-(2-hydroxyethyl)-5-vinylnicotinic acid gamma-lactone; 5-Ethenyl-3,4-dihydro-1H-pyrano(3,4-c)pyridin-1-one; 1H-Pyrano(3,4-c)pyridin-1-one, 5-ethenyl-3,4-dihydro-; Erythricine; UNII-C2PD310UXB; Gencianina; BRN 0137011; GENTIANINE [MI]; C06525; 4-27-00-02817 (Beilstein Handbook Reference); SCHEMBL2216852; CHEMBL4753058; CHEBI:28981; DTXSID00963141; Nicotinic acid, 4-(2-hydroxyethyl)-5-vinyl-, gamma-lactone; HY-N6039; ZINC1530467; BBL036668; STL559047; AKOS005266580; VS-13618; CS-0032220; From Schultesia guianensis malme (Mata-Zombando); Q-100479; Q27104004; 1H-Pyrano(3,4-c)pyridin-1-one, 3,4-dihydro-5-vinyl-; 5-VINYL-3,4-DIHYDROPYRANO(3,4-C)PYRIDIN-1-ONE; NICOTINIC ACID, 4-(2-HYDROXYETHYL)-5-VINYL-, .DELTA.-LACTONE
|
|
| CAS | 439-89-4 | |
| PubChem CID | 354616 | |
| ChEMBL ID | CHEMBL4753058 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 175.18 | ALogp: | 1.5 |
| HBD: | 0 | HBA: | 3 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 39.2 | Aromatic Rings: | 2 |
| Heavy Atoms: | 13 | QED Weighted: | 0.612 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.661 | MDCK Permeability: | 0.00003450 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.084 |
| 30% Bioavailability (F30%): | 0.441 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.904 | Plasma Protein Binding (PPB): | 70.27% |
| Volume Distribution (VD): | 1.786 | Fu: | 35.81% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.9 | CYP1A2-substrate: | 0.463 |
| CYP2C19-inhibitor: | 0.332 | CYP2C19-substrate: | 0.507 |
| CYP2C9-inhibitor: | 0.118 | CYP2C9-substrate: | 0.695 |
| CYP2D6-inhibitor: | 0.03 | CYP2D6-substrate: | 0.801 |
| CYP3A4-inhibitor: | 0.565 | CYP3A4-substrate: | 0.285 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 12.687 | Half-life (T1/2): | 0.718 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.046 | Human Hepatotoxicity (H-HT): | 0.141 |
| Drug-inuced Liver Injury (DILI): | 0.408 | AMES Toxicity: | 0.036 |
| Rat Oral Acute Toxicity: | 0.338 | Maximum Recommended Daily Dose: | 0.303 |
| Skin Sensitization: | 0.853 | Carcinogencity: | 0.537 |
| Eye Corrosion: | 0.025 | Eye Irritation: | 0.638 |
| Respiratory Toxicity: | 0.179 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC006137 | 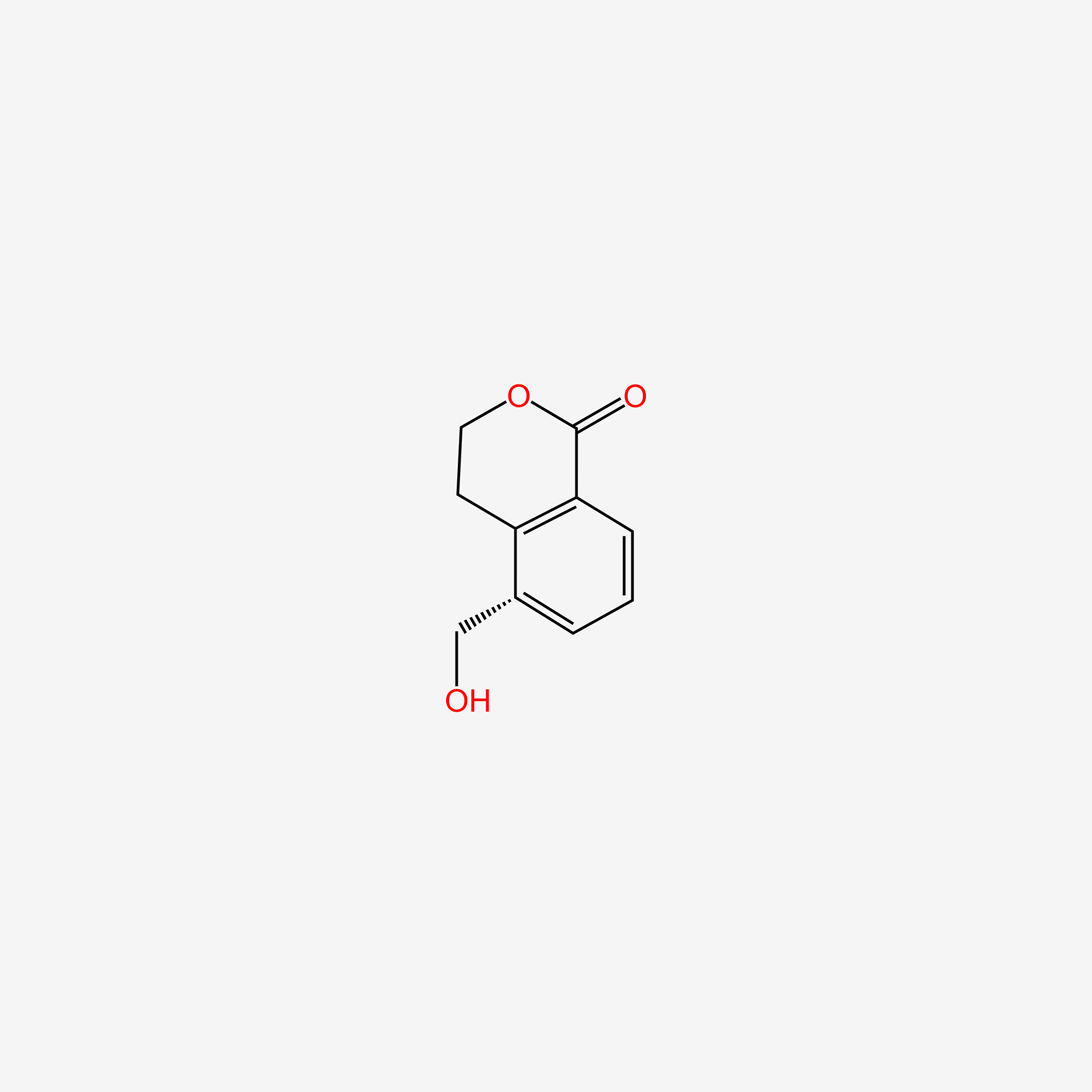 |
0.396 | D0Z8AA | 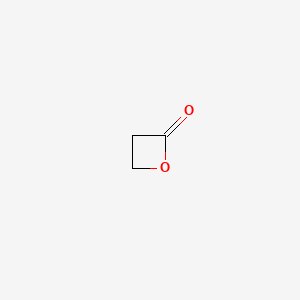 |
0.256 | ||
| ENC001074 | 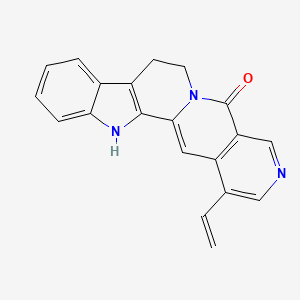 |
0.289 | D0U0KW | 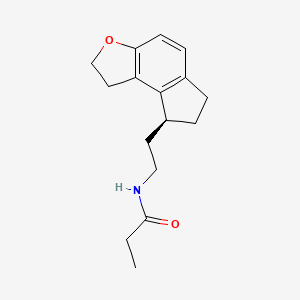 |
0.211 | ||
| ENC002244 | 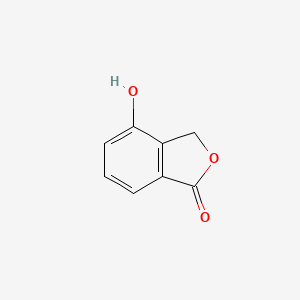 |
0.283 | D0Q5MQ | 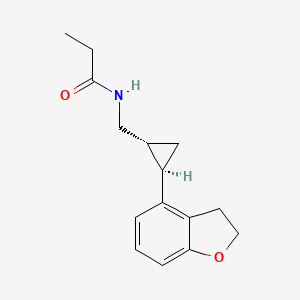 |
0.203 | ||
| ENC006136 | 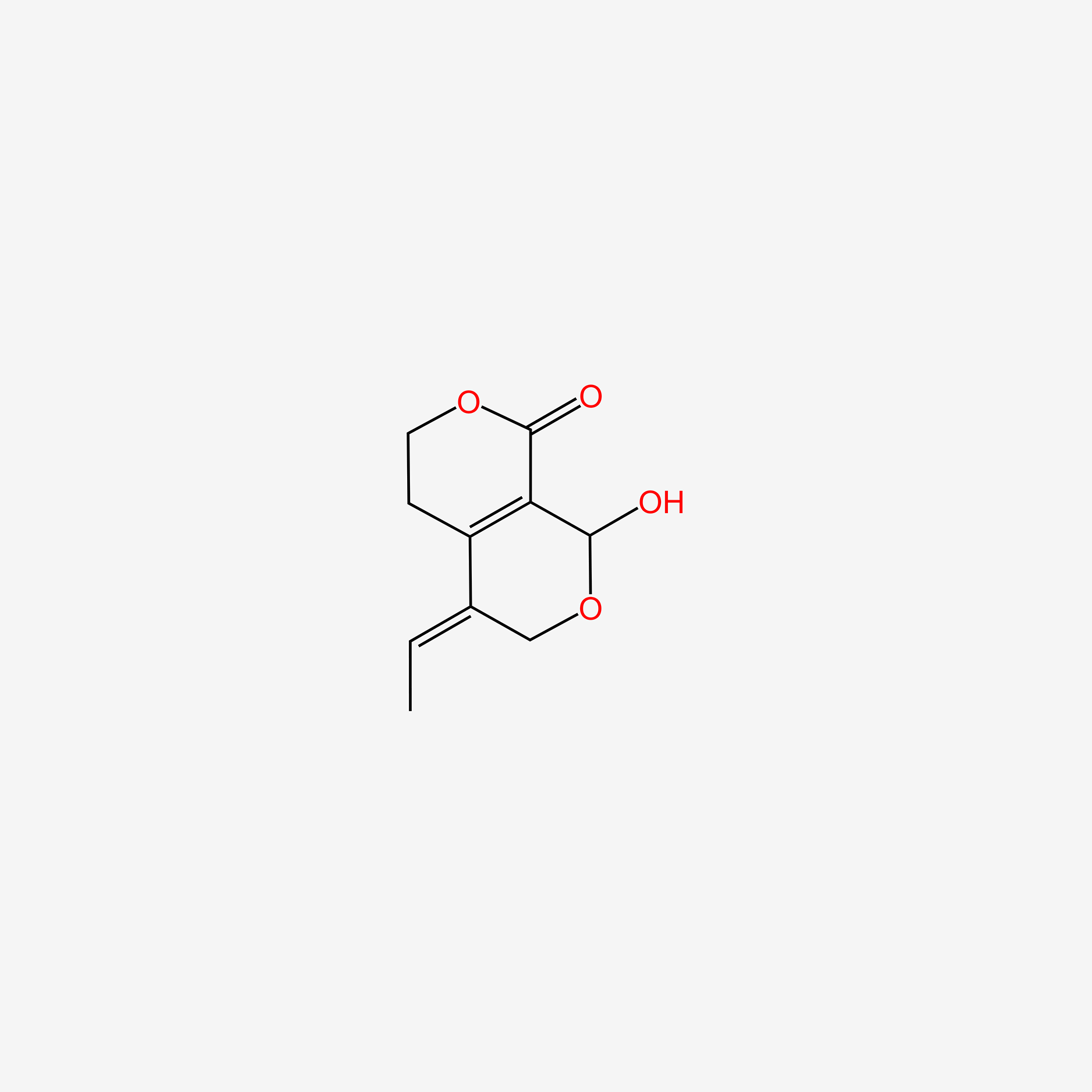 |
0.267 | D0TY5N | 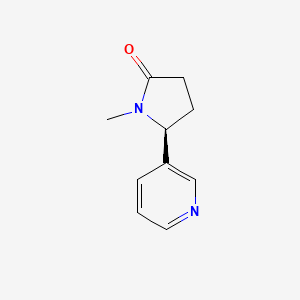 |
0.194 | ||
| ENC005108 | 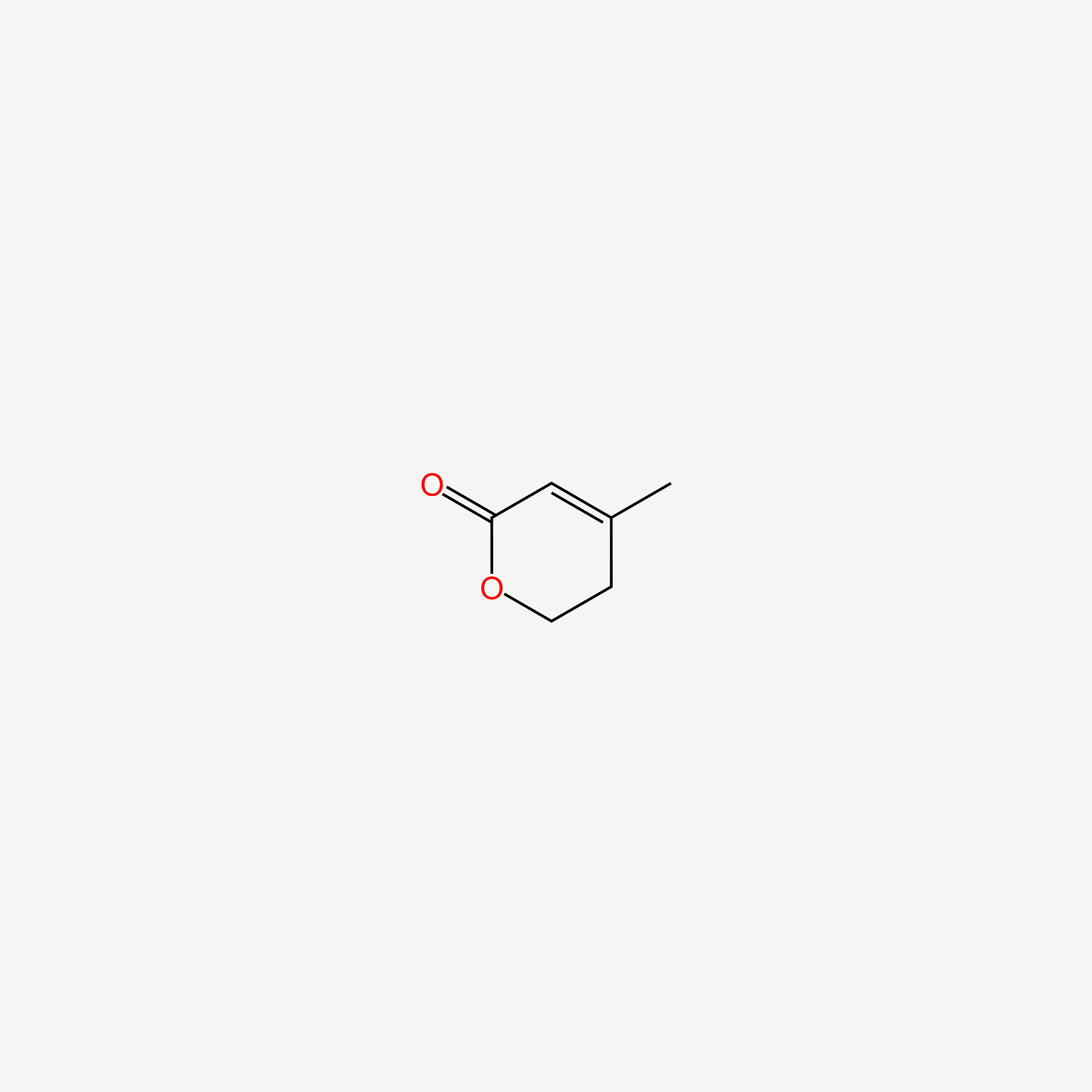 |
0.255 | D03CUF | 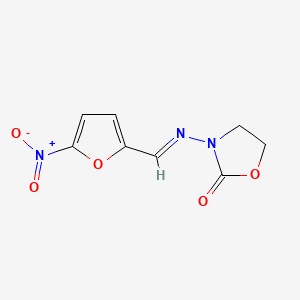 |
0.188 | ||
| ENC001280 | 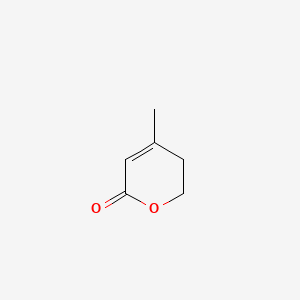 |
0.255 | D0Y8ZN | 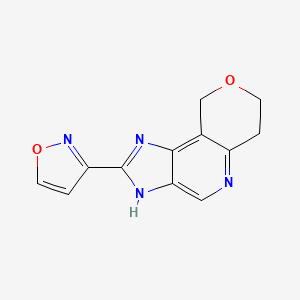 |
0.182 | ||
| ENC002321 | 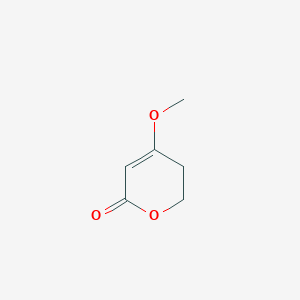 |
0.240 | D06HLY | 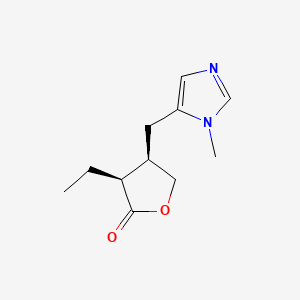 |
0.179 | ||
| ENC005295 | 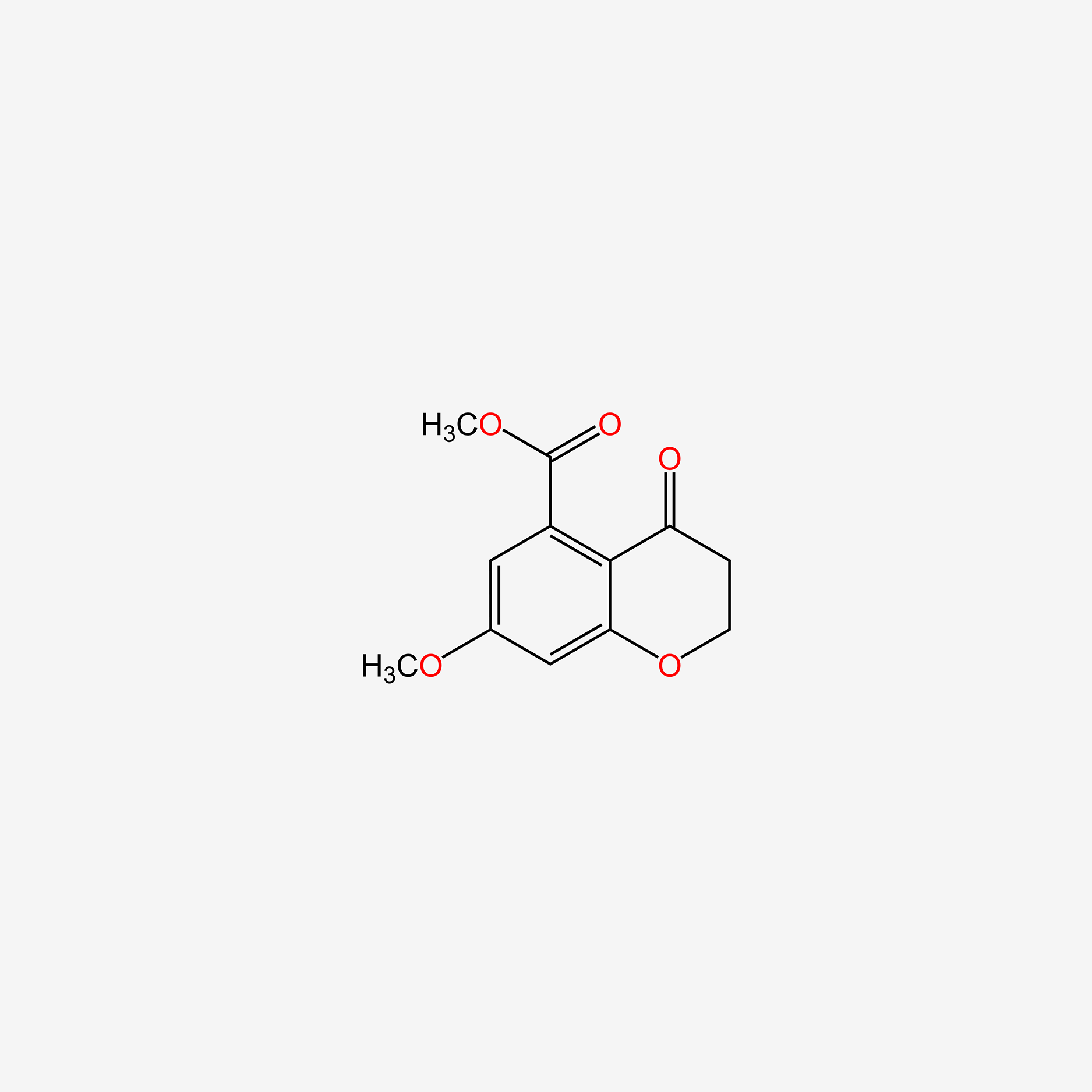 |
0.217 | D00EEL | 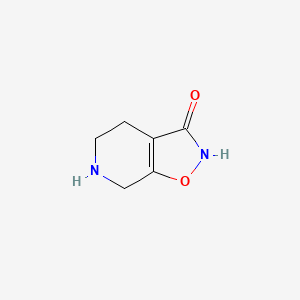 |
0.179 | ||
| ENC000184 | 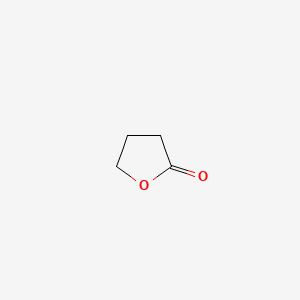 |
0.200 | D0D1HW | 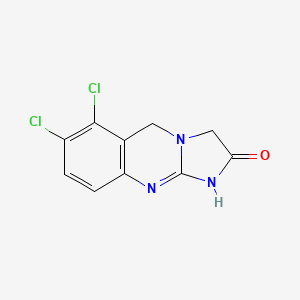 |
0.171 | ||
| ENC005453 | 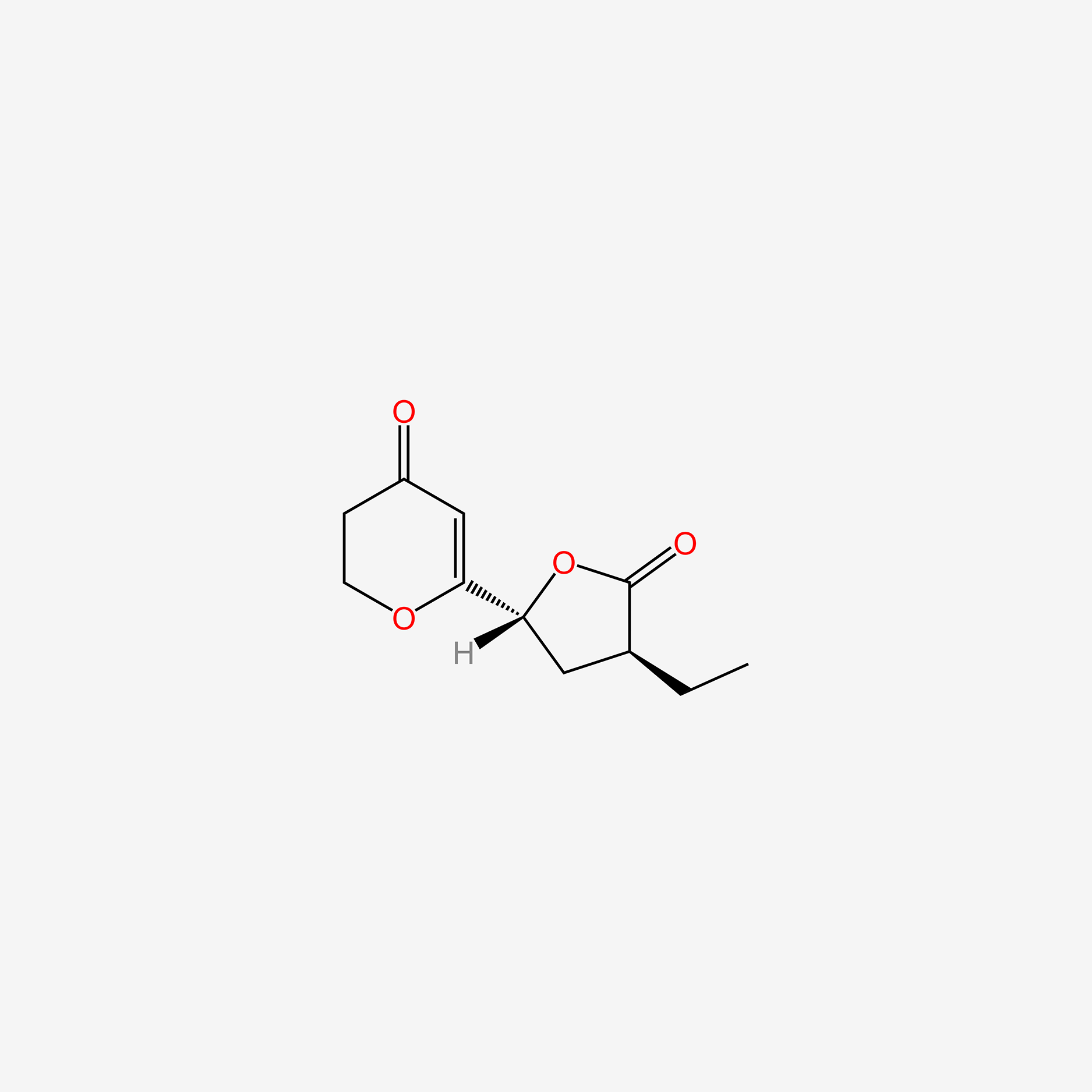 |
0.197 | D0N1WU | 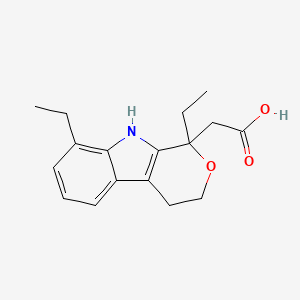 |
0.171 | ||