NPs Basic Information
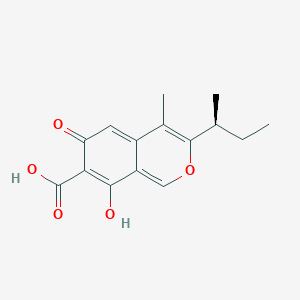
|
Name |
Ascochitine
|
| Molecular Formula | C15H16O5 | |
| IUPAC Name* |
3-[(2S)-butan-2-yl]-8-hydroxy-4-methyl-6-oxoisochromene-7-carboxylic acid
|
|
| SMILES |
CC[C@H](C)C1=C(C2=CC(=O)C(=C(C2=CO1)O)C(=O)O)C
|
|
| InChI |
InChI=1S/C15H16O5/c1-4-7(2)14-8(3)9-5-11(16)12(15(18)19)13(17)10(9)6-20-14/h5-7,17H,4H2,1-3H3,(H,18,19)/t7-/m0/s1
|
|
| InChIKey |
DZBCEUTUWOWUDY-ZETCQYMHSA-N
|
|
| Synonyms |
Ascochitine; Ascochitin; 3615-05-2; NSC114344; CO17L3VWWJ; Aschchitine from B624868 K051; NSC-114344; Ascochytin; 6H-2-Benzopyran-7-carboxylic acid, 8-hydroxy-4-methyl-3-(1-methylpropyl)-6-oxo-, (S)-; 6H-2-Benzopyran-7-carboxylic acid, 8-hydroxy-4-methyl-3-[(1S)-1-methylpropyl]-6-oxo-; 6H-2-BENZOPYRAN-7-CARBOXYLIC ACID, 8-HYDROXY-4-METHYL-3-((1S)-1-METHYLPROPYL)-6-OXO-; UNII-CO17L3VWWJ; CHEMBL1964740; DTXSID80957563; CHEBI:172513; ZINC1704485; NSC 114344; NCI60_000318; Q27275562; 3-[(2S)-butan-2-yl]-8-hydroxy-4-methyl-6-oxoisochromene-7-carboxylic acid; 3-(Butan-2-yl)-8-hydroxy-4-methyl-6-oxo-6H-2-benzopyran-7-carboxylic acid; 6H-2-BENZOPYRAN-7-CARBOXYLIC ACID, 3-SEC-BUTYL-8-HYDROXY-4-METHYL-6-OXO-, (S)-(-)-
|
|
| CAS | 3615-05-2 | |
| PubChem CID | 73486 | |
| ChEMBL ID | CHEMBL1964740 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 276.28 | ALogp: | 2.1 |
| HBD: | 2 | HBA: | 5 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 83.8 | Aromatic Rings: | 2 |
| Heavy Atoms: | 20 | QED Weighted: | 0.891 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.09 | MDCK Permeability: | 0.00000709 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.175 |
| Human Intestinal Absorption (HIA): | 0.025 | 20% Bioavailability (F20%): | 0.031 |
| 30% Bioavailability (F30%): | 0.003 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.035 | Plasma Protein Binding (PPB): | 95.29% |
| Volume Distribution (VD): | 0.973 | Fu: | 5.89% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.741 | CYP1A2-substrate: | 0.573 |
| CYP2C19-inhibitor: | 0.105 | CYP2C19-substrate: | 0.057 |
| CYP2C9-inhibitor: | 0.578 | CYP2C9-substrate: | 0.124 |
| CYP2D6-inhibitor: | 0.743 | CYP2D6-substrate: | 0.128 |
| CYP3A4-inhibitor: | 0.103 | CYP3A4-substrate: | 0.095 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 1.162 | Half-life (T1/2): | 0.895 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.017 | Human Hepatotoxicity (H-HT): | 0.416 |
| Drug-inuced Liver Injury (DILI): | 0.975 | AMES Toxicity: | 0.022 |
| Rat Oral Acute Toxicity: | 0.425 | Maximum Recommended Daily Dose: | 0.245 |
| Skin Sensitization: | 0.22 | Carcinogencity: | 0.711 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.32 |
| Respiratory Toxicity: | 0.93 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005371 | 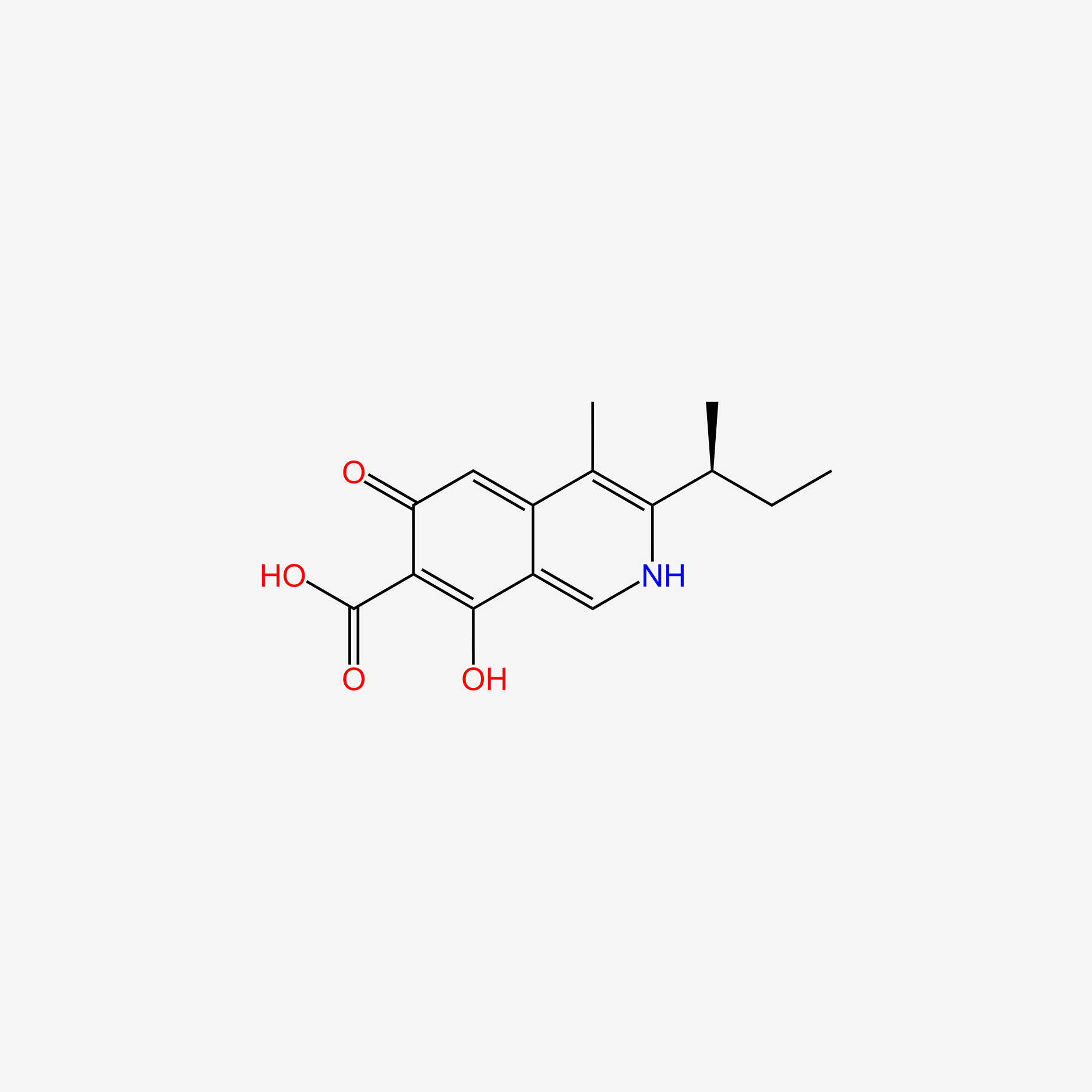 |
0.683 | D0G5UB | 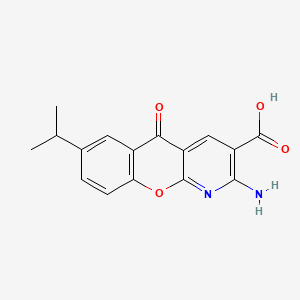 |
0.256 | ||
| ENC003256 | 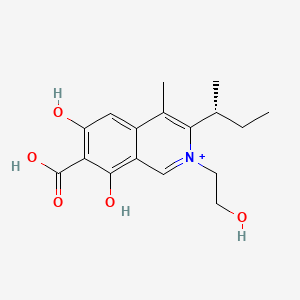 |
0.407 | D07JGT |  |
0.253 | ||
| ENC005100 | 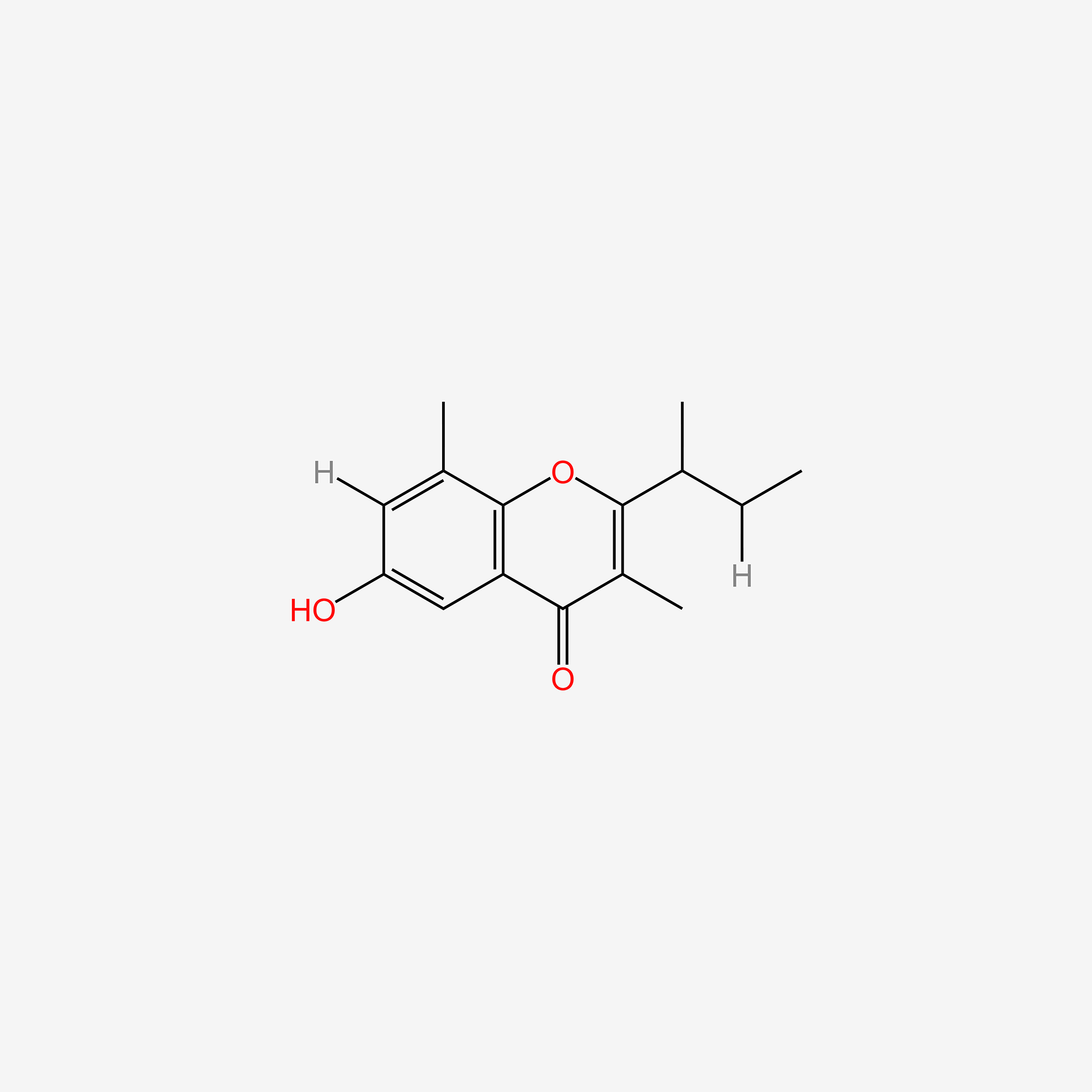 |
0.403 | D0O6KE | 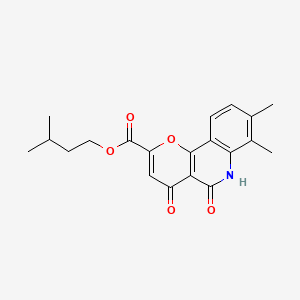 |
0.253 | ||
| ENC002605 | 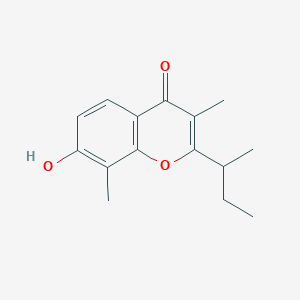 |
0.384 | D06FVX | 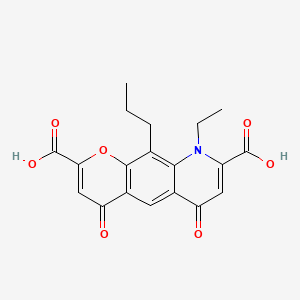 |
0.248 | ||
| ENC005367 |  |
0.364 | D07UXP | 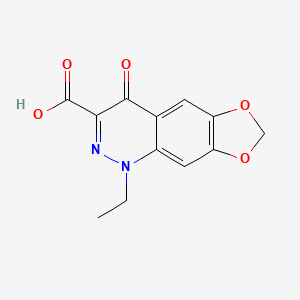 |
0.247 | ||
| ENC006097 | 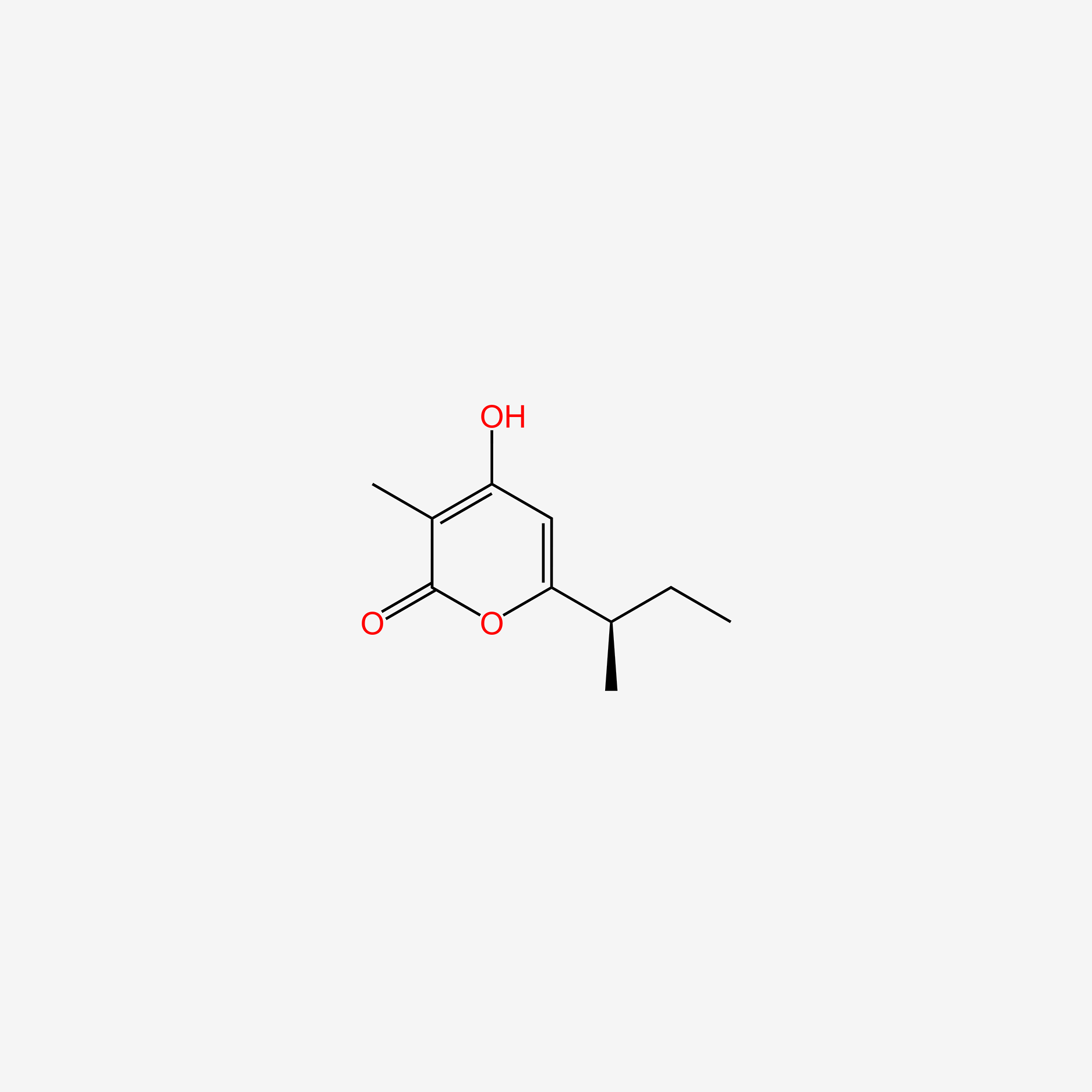 |
0.359 | D0A5SE | 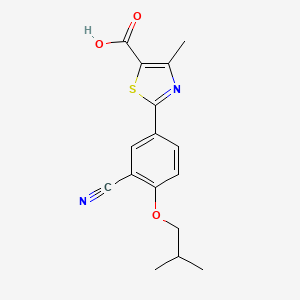 |
0.242 | ||
| ENC002326 | 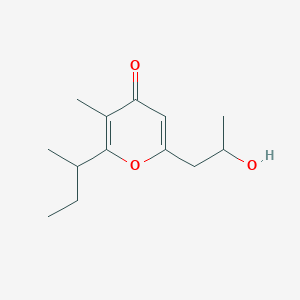 |
0.357 | D0WY9N | 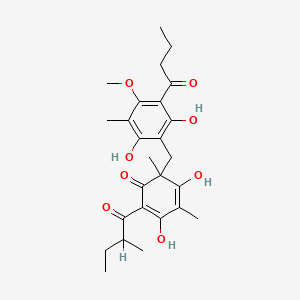 |
0.237 | ||
| ENC004131 | 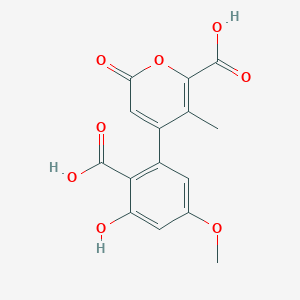 |
0.357 | D0Z3DY | 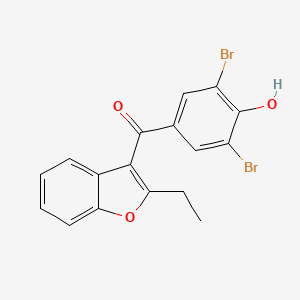 |
0.226 | ||
| ENC001445 | 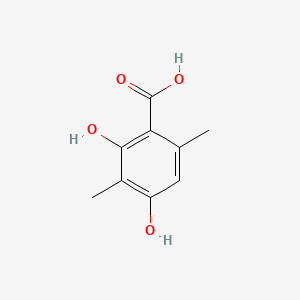 |
0.344 | D0QD1G | 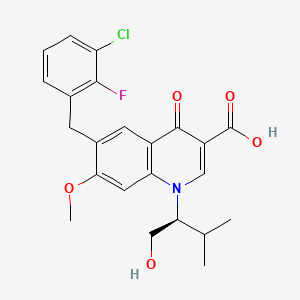 |
0.223 | ||
| ENC000945 | 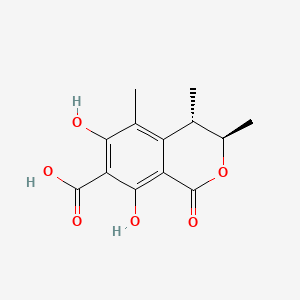 |
0.342 | D0Z1WA | 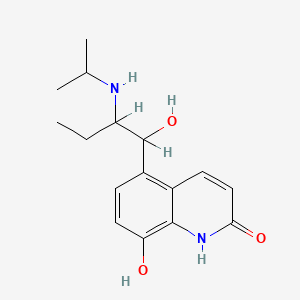 |
0.222 | ||