NPs Basic Information
|
Name |
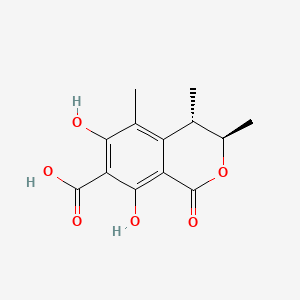
Dihydrocitrinone
|
| Molecular Formula | C13H14O6 | |
| IUPAC Name* |
(3R,4S)-6,8-dihydroxy-3,4,5-trimethyl-1-oxo-3,4-dihydroisochromene-7-carboxylic acid
|
|
| SMILES |
C[C@@H]1[C@H](OC(=O)C2=C(C(=C(C(=C12)C)O)C(=O)O)O)C
|
|
| InChI |
InChI=1S/C13H14O6/c1-4-6(3)19-13(18)8-7(4)5(2)10(14)9(11(8)15)12(16)17/h4,6,14-15H,1-3H3,(H,16,17)/t4-,6-/m1/s1
|
|
| InChIKey |
VVVMDYGNIVXIIG-INEUFUBQSA-N
|
|
| Synonyms |
Dihydrocitrinone; 65718-85-6; (3R,4S)-6,8-dihydroxy-3,4,5-trimethyl-1-oxo-3,4-dihydroisochromene-7-carboxylic acid; 9I8K50016V; 1H-2-Benzopyran-7-carboxylic acid, 3,4-dihydro-6,8-dihydroxy-3,4,5-trimethyl-1-oxo-, (3R-trans)-; UNII-9I8K50016V; CITRINONE, DIHYDRO-; CHEMBL465858; MEGxm0_000092; ACon0_000322; ACon1_001305; DTXSID10215926; ZINC6069169; NCGC00180648-01; BRD-K21191814-001-01-1; Q27272591; (3R,4S)-3,4-DIHYDRO-6,8-DIHYDROXY-3,4,5-TRIMETHYL-1-OXO-1H-2-BENZOPYRAN-7-CARBOXYLIC ACID; 1H-2-BENZOPYRAN-7-CARBOXYLIC ACID, 3,4-DIHYDRO-6,8-DIHYDROXY-3,4,5-TRIMETHYL-1-OXO-, (3R,4S)-; NCGC00180648-02!(3R,4S)-6,8-dihydroxy-3,4,5-trimethyl-1-oxo-3,4-dihydroisochromene-7-carboxylic acid
|
|
| CAS | 65718-85-6 | |
| PubChem CID | 163095 | |
| ChEMBL ID | CHEMBL465858 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 266.25 | ALogp: | 2.9 |
| HBD: | 3 | HBA: | 6 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 104.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 19 | QED Weighted: | 0.674 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.743 | MDCK Permeability: | 0.00000658 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.026 | 20% Bioavailability (F20%): | 0.011 |
| 30% Bioavailability (F30%): | 0.023 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.056 | Plasma Protein Binding (PPB): | 97.84% |
| Volume Distribution (VD): | 0.425 | Fu: | 2.34% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.119 | CYP1A2-substrate: | 0.436 |
| CYP2C19-inhibitor: | 0.033 | CYP2C19-substrate: | 0.057 |
| CYP2C9-inhibitor: | 0.281 | CYP2C9-substrate: | 0.109 |
| CYP2D6-inhibitor: | 0.083 | CYP2D6-substrate: | 0.102 |
| CYP3A4-inhibitor: | 0.062 | CYP3A4-substrate: | 0.056 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 1.16 | Half-life (T1/2): | 0.745 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.004 | Human Hepatotoxicity (H-HT): | 0.273 |
| Drug-inuced Liver Injury (DILI): | 0.97 | AMES Toxicity: | 0.014 |
| Rat Oral Acute Toxicity: | 0.07 | Maximum Recommended Daily Dose: | 0.022 |
| Skin Sensitization: | 0.111 | Carcinogencity: | 0.085 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.409 |
| Respiratory Toxicity: | 0.347 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC004924 | 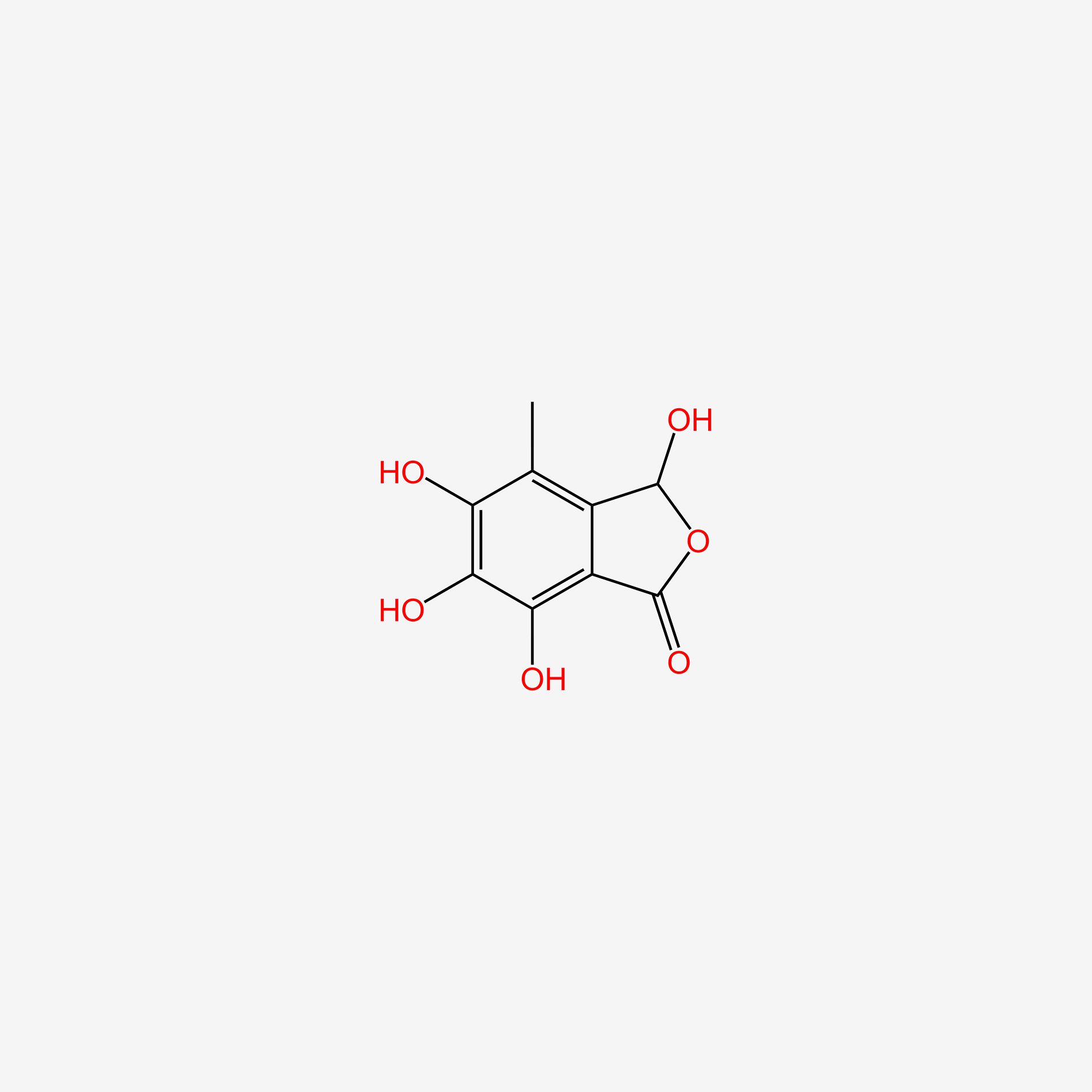 |
0.467 | D01XWG | 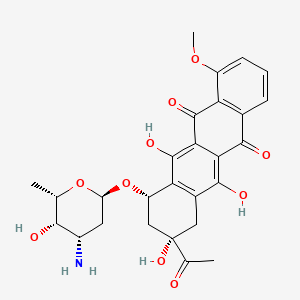 |
0.236 | ||
| ENC002391 | 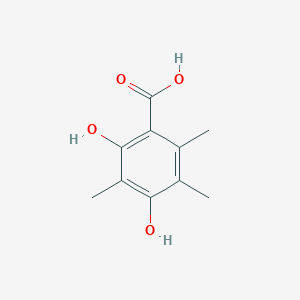 |
0.448 | D01XDL | 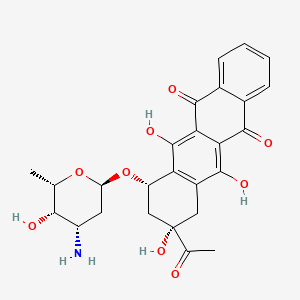 |
0.235 | ||
| ENC005912 | 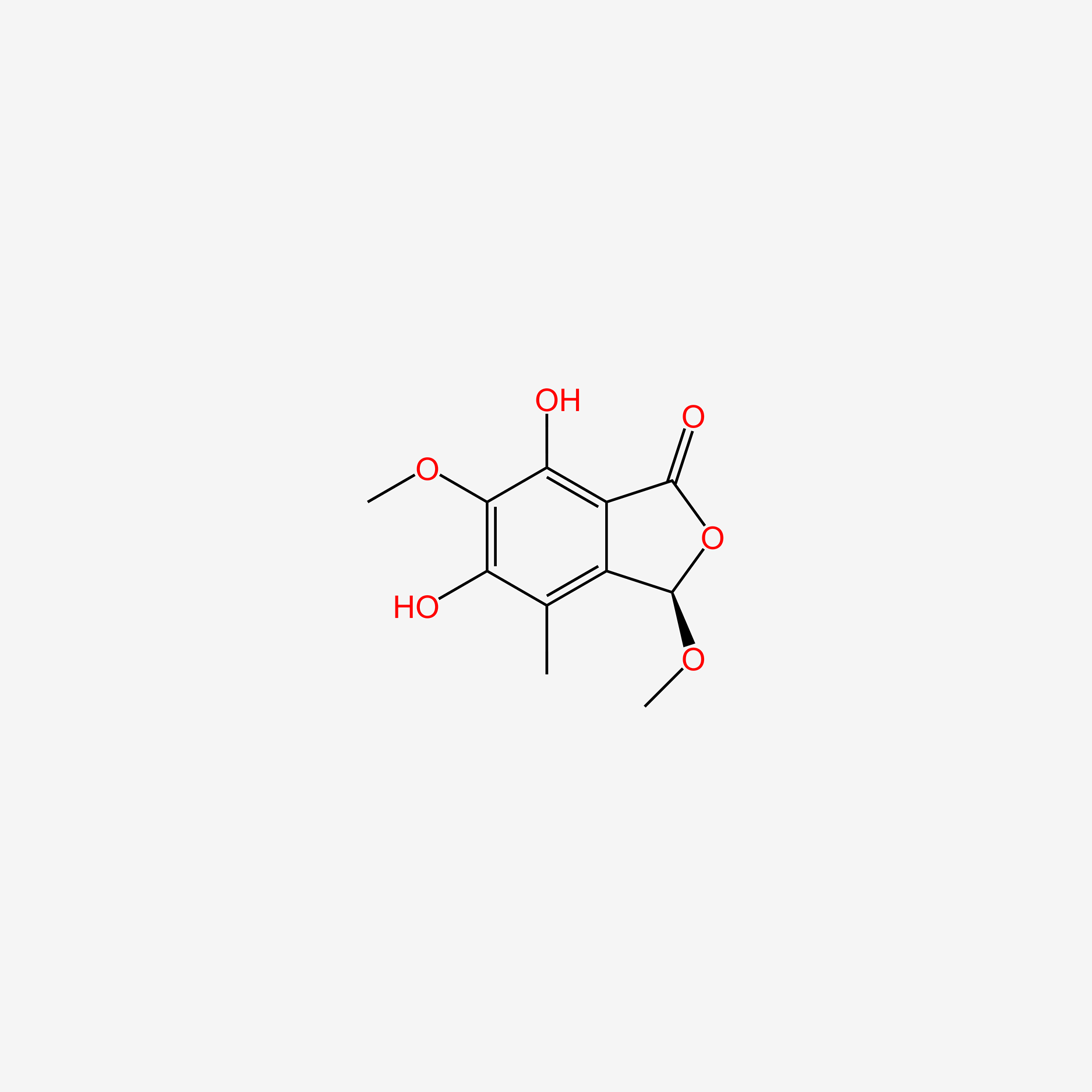 |
0.446 | D0S0LZ |  |
0.231 | ||
| ENC005911 | 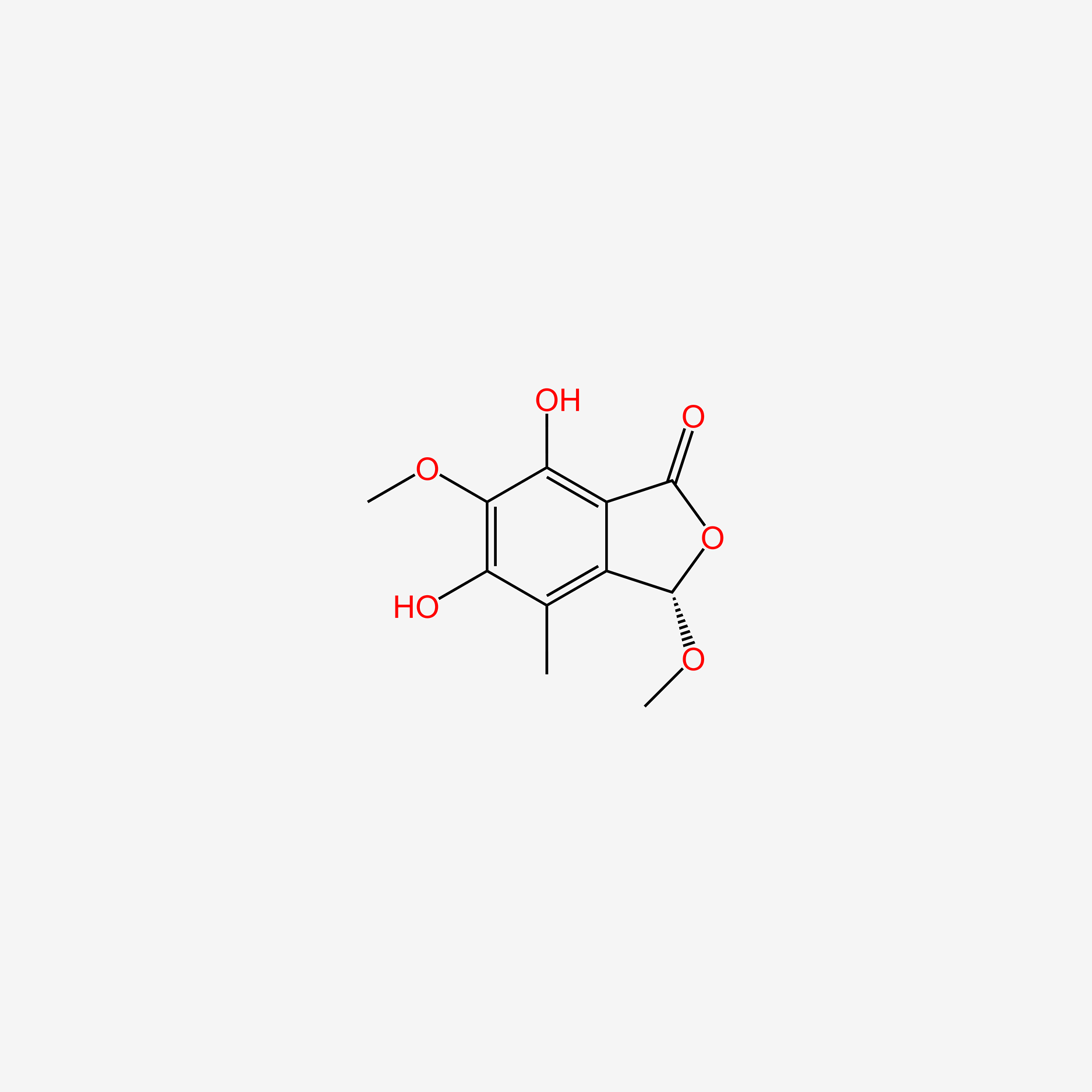 |
0.446 | D0JL2K | 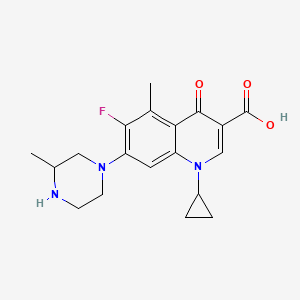 |
0.224 | ||
| ENC004367 | 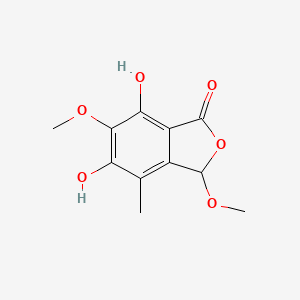 |
0.446 | D0WY9N | 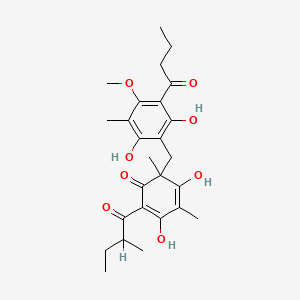 |
0.223 | ||
| ENC004991 | 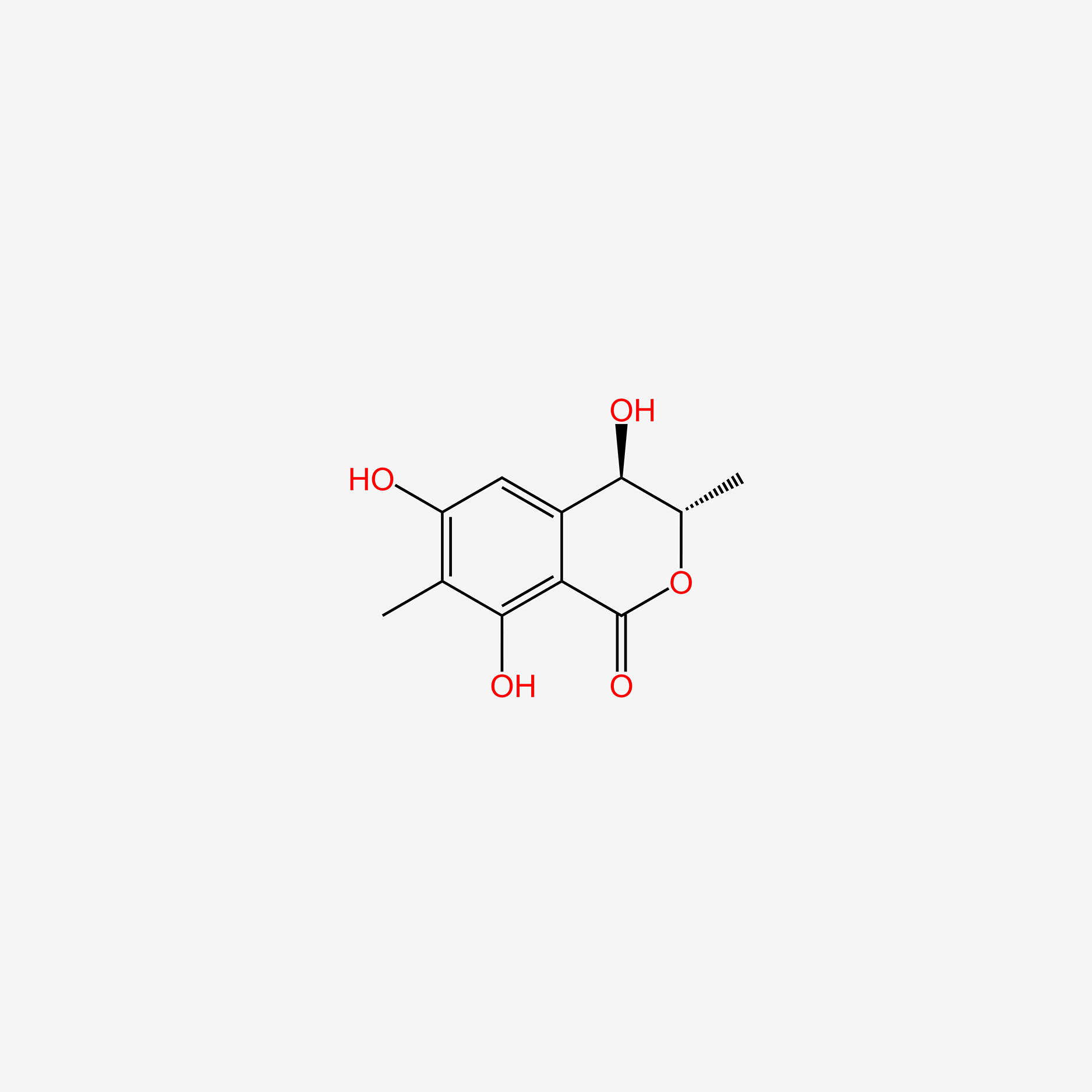 |
0.444 | D0YH0N | 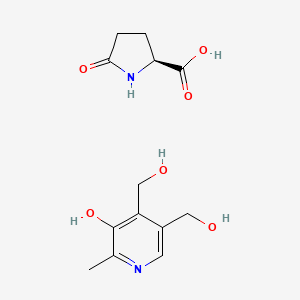 |
0.221 | ||
| ENC004363 |  |
0.444 | D07VLY |  |
0.220 | ||
| ENC003148 |  |
0.439 | D0C9XJ |  |
0.220 | ||
| ENC003702 | 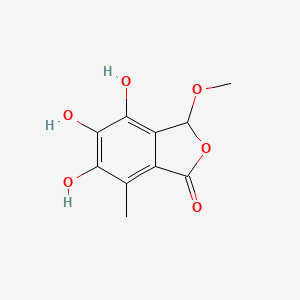 |
0.422 | D0Q2AT | 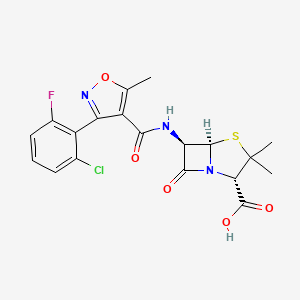 |
0.215 | ||
| ENC004969 | 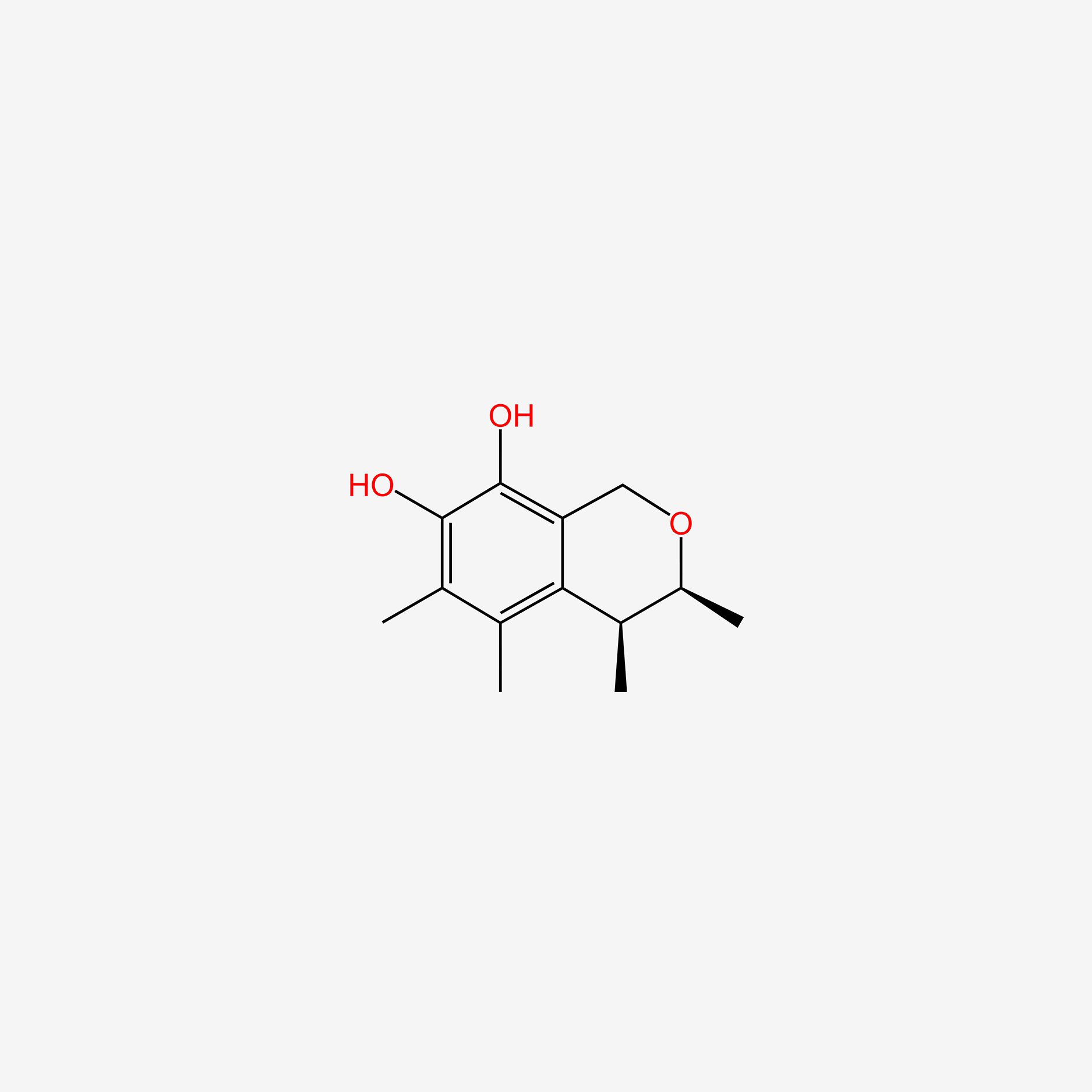 |
0.400 | D0R2KJ | 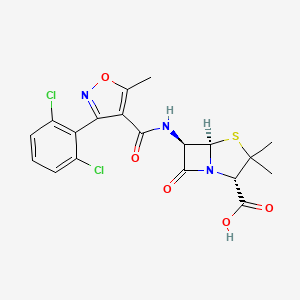 |
0.215 | ||