NPs Basic Information
|
Name |
beta-Phellandrene
|
| Molecular Formula | C10H16 | |
| IUPAC Name* |
3-methylidene-6-propan-2-ylcyclohexene
|
|
| SMILES |
CC(C)C1CCC(=C)C=C1
|
|
| InChI |
InChI=1S/C10H16/c1-8(2)10-6-4-9(3)5-7-10/h4,6,8,10H,3,5,7H2,1-2H3
|
|
| InChIKey |
LFJQCDVYDGGFCH-UHFFFAOYSA-N
|
|
| Synonyms |
BETA-PHELLANDRENE; 555-10-2; p-Mentha-1(7),2-diene; 2-p-Menthadiene; 3-Isopropyl-6-methylenecyclohex-1-ene; .beta.-Phellandrene; Cyclohexene, 3-methylene-6-(1-methylethyl)-; 3-Methylene-6-(1-methylethyl)cyclohexene; PHELLANDRENE, BETA; 3-Isopropyl-6-methylene-1-cyclohexene; 4-Isopropyl-1-methylene-2-cyclohexene; 3-methylidene-6-propan-2-ylcyclohexene; beta-Phellandren; CHEBI:48741; 3-methylidene-6-(propan-2-yl)cyclohex-1-ene; NSC-53044; 2KK225M001; b-phellandrene; UNII-2KK225M001; HSDB 4080; beta -phellandrene; EINECS 209-081-9; NSC 53044; Phellandrene, .beta.; Epitope ID:123895; 3-METHYLIDENE-6-PROPAN-2-YL-CYCLOHEXENE; CHEMBL444254; BETA-PHELLANDRENE [HSDB]; DTXSID4052215; .BETA.-PHELLANDRENE [MI]; 3-Isopropyl-6-methylenecyclohexene; HY-N8573; NSC53044; (+/-)-.BETA.-PHELLANDRENE; AKOS016008994; 3-Isopropyl-6-methylene-1-cyclohexene #; 3-Methylene-6-(1-methylethyl)-Cyclohexene; 3-methylene-6-(1-methylethenyl)-cyclohexane; CS-0148637; C19818; EN300-2528048; Q19606727
|
|
| CAS | 555-10-2 | |
| PubChem CID | 11142 | |
| ChEMBL ID | CHEMBL444254 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 136.23 | ALogp: | 3.4 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.512 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.382 | MDCK Permeability: | 0.00002850 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.003 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.969 | Plasma Protein Binding (PPB): | 89.57% |
| Volume Distribution (VD): | 2.149 | Fu: | 8.47% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.409 | CYP1A2-substrate: | 0.38 |
| CYP2C19-inhibitor: | 0.142 | CYP2C19-substrate: | 0.866 |
| CYP2C9-inhibitor: | 0.211 | CYP2C9-substrate: | 0.347 |
| CYP2D6-inhibitor: | 0.012 | CYP2D6-substrate: | 0.653 |
| CYP3A4-inhibitor: | 0.166 | CYP3A4-substrate: | 0.567 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.289 | Half-life (T1/2): | 0.479 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.031 | Human Hepatotoxicity (H-HT): | 0.269 |
| Drug-inuced Liver Injury (DILI): | 0.192 | AMES Toxicity: | 0.024 |
| Rat Oral Acute Toxicity: | 0.045 | Maximum Recommended Daily Dose: | 0.83 |
| Skin Sensitization: | 0.93 | Carcinogencity: | 0.874 |
| Eye Corrosion: | 0.3 | Eye Irritation: | 0.906 |
| Respiratory Toxicity: | 0.911 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
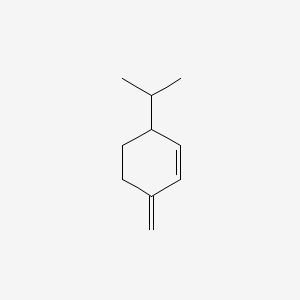
| ENC002234 | 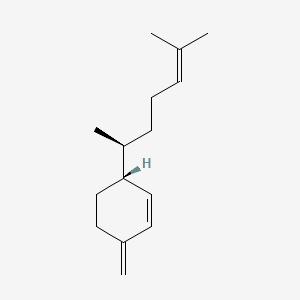 |
0.511 | D04CSZ | 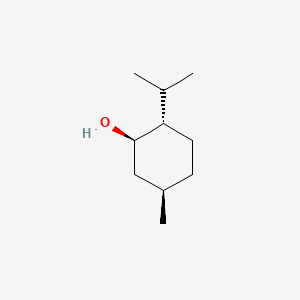 |
0.217 | ||
| ENC000196 | 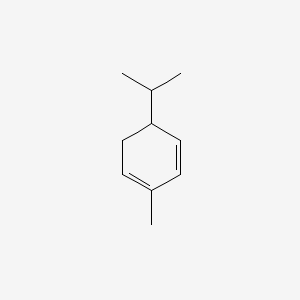 |
0.421 | D0P4MT | 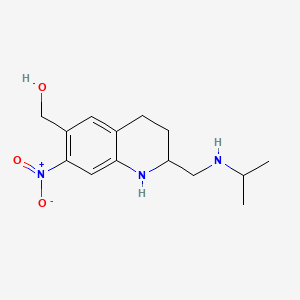 |
0.188 | ||
| ENC000872 | 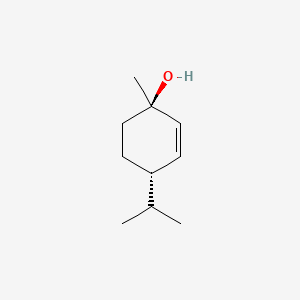 |
0.400 | D0Z8SF | 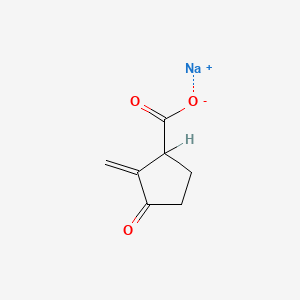 |
0.174 | ||
| ENC002264 | 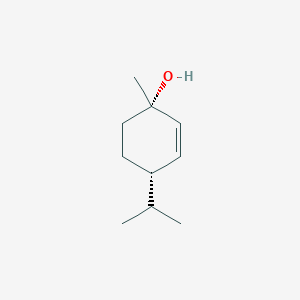 |
0.400 | D0D2VS | 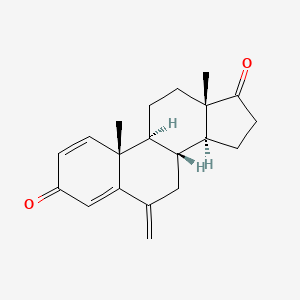 |
0.173 | ||
| ENC001809 | 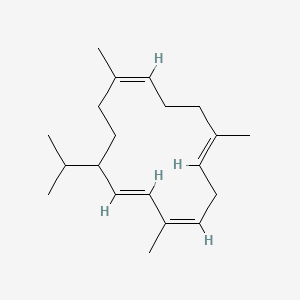 |
0.344 | D06PSS | 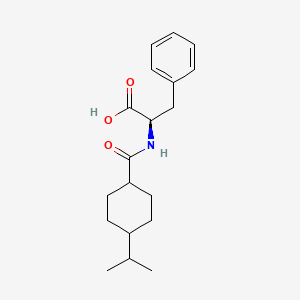 |
0.167 | ||
| ENC003463 | 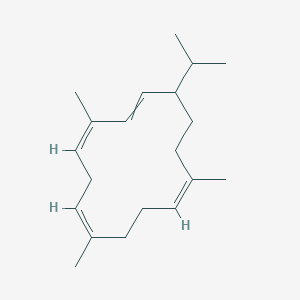 |
0.344 | D06GIP | 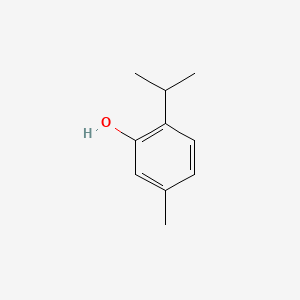 |
0.167 | ||
| ENC003090 | 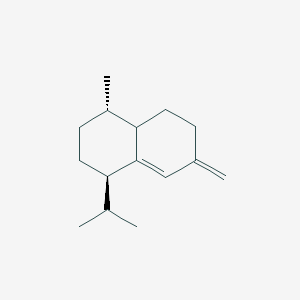 |
0.333 | D0TY5N | 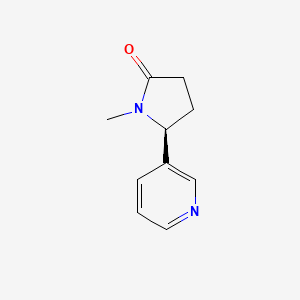 |
0.164 | ||
| ENC000165 | 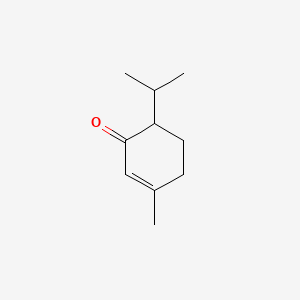 |
0.302 | D03DVJ | 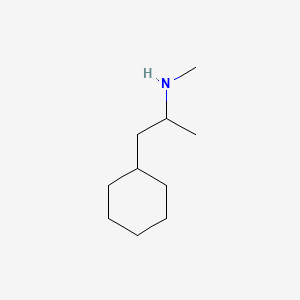 |
0.160 | ||
| ENC000763 | 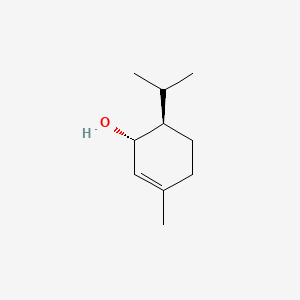 |
0.302 | D01CKY | 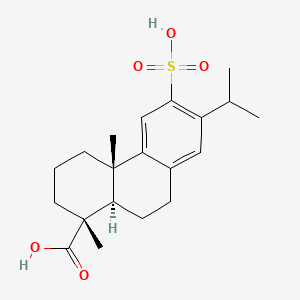 |
0.157 | ||
| ENC000762 | 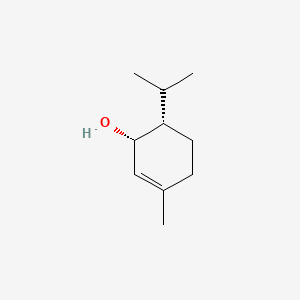 |
0.302 | D0A6CQ | 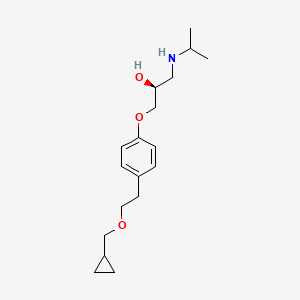 |
0.156 | ||