NPs Basic Information
|
Name |
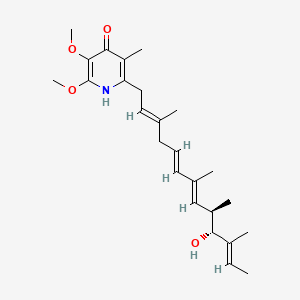
Piericidin A
|
| Molecular Formula | C25H37NO4 | |
| IUPAC Name* |
2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraenyl]-5,6-dimethoxy-3-methyl-1H-pyridin-4-one
|
|
| SMILES |
C/C=C(\C)/[C@@H]([C@H](C)/C=C(\C)/C=C/C/C(=C/CC1=C(C(=O)C(=C(N1)OC)OC)C)/C)O
|
|
| InChI |
InChI=1S/C25H37NO4/c1-9-18(4)22(27)19(5)15-17(3)12-10-11-16(2)13-14-21-20(6)23(28)24(29-7)25(26-21)30-8/h9-10,12-13,15,19,22,27H,11,14H2,1-8H3,(H,26,28)/b12-10+,16-13+,17-15+,18-9+/t19-,22+/m1/s1
|
|
| InChIKey |
BBLGCDSLCDDALX-LKGBESRRSA-N
|
|
| Synonyms |
Piericidin A; Piericidin A1; Shaoguanmycin B; 2738-64-9; Piericidine A; SN 198E; 8VT513UJ9R; CHEMBL272733; 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraenyl]-5,6-dimethoxy-3-methyl-1H-pyridin-4-one; Piericidin; BRN 1555726; UNII-8VT513UJ9R; AR 054; PIERICIDINA; (+)-piericidin A1; 4-Pyridinol, 2-(10-hydroxy-3,7,9,11-tetramethyl-2,5,7,11-tridecatetraenyl)-5,6-dimethoxy-3-methyl-, (R-(R*,R*-(all-E)))-; MEGxm0_000313; PIERICIDIN A, (+)-; SCHEMBL18941698; SCHEMBL19717606; ACon0_001227; ACon1_001455; DTXSID80880044; SN-198E; CHEBI:138511; HQH; BDBM50411905; ZINC14655907; NCGC00180489-01; 2,6,9,11-Tridecatetraen-4-ol, 13-(4-hydroxy-5,6-dimethoxy-3-methyl-2-pyridyl)-3,5,7,11-tetramethyl-, (all-E)-(4R,5R)-; 4-Pyridinol, 2-((2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyl-2,5,7,11-tridecatetraenyl)-5,6-dimethoxy-3-methyl-; HY-114936; CS-0064672; J-016753; Q7191888; BRD-K73581776-001-01-6; Piericidin A from Streptomyces mobaraensis, >=90.0% (HPLC), liquid, green-yellow; 19855-42-6; 2-[(2E,5E,7E,11E)-10R-hydroxy-3,7,9R,11-tetramethyl-2,5,7,11-tridecatetraen-1-yl]-5,6-dimethoxy-3-methyl-4-pyridinol; 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraen-1-yl]-5,6-dimethoxy-3-methylpyridin-4-ol; 4-PYRIDINOL, 2-((2E,5E,7E,9R,10R,11E)-10-HYDROXY-3,7,9,11-TETRAMETHYL-2,5,7,11-TRIDECATETRAEN-1-YL)-5,6-DIMETHOXY-3-METHYL-
|
|
| CAS | 2738-64-9 | |
| PubChem CID | 6437838 | |
| ChEMBL ID | CHEMBL272733 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 415.6 | ALogp: | 6.1 |
| HBD: | 2 | HBA: | 5 |
| Rotatable Bonds: | 10 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 67.8 | Aromatic Rings: | 1 |
| Heavy Atoms: | 30 | QED Weighted: | 0.398 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.655 | MDCK Permeability: | 0.00001230 |
| Pgp-inhibitor: | 0.018 | Pgp-substrate: | 0.546 |
| Human Intestinal Absorption (HIA): | 0.027 | 20% Bioavailability (F20%): | 0.026 |
| 30% Bioavailability (F30%): | 0.024 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.033 | Plasma Protein Binding (PPB): | 96.93% |
| Volume Distribution (VD): | 0.366 | Fu: | 1.62% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.389 | CYP1A2-substrate: | 0.954 |
| CYP2C19-inhibitor: | 0.738 | CYP2C19-substrate: | 0.87 |
| CYP2C9-inhibitor: | 0.725 | CYP2C9-substrate: | 0.967 |
| CYP2D6-inhibitor: | 0.896 | CYP2D6-substrate: | 0.899 |
| CYP3A4-inhibitor: | 0.914 | CYP3A4-substrate: | 0.752 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 3.943 | Half-life (T1/2): | 0.553 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.019 | Human Hepatotoxicity (H-HT): | 0.184 |
| Drug-inuced Liver Injury (DILI): | 0.547 | AMES Toxicity: | 0.005 |
| Rat Oral Acute Toxicity: | 0.026 | Maximum Recommended Daily Dose: | 0.839 |
| Skin Sensitization: | 0.948 | Carcinogencity: | 0.017 |
| Eye Corrosion: | 0.004 | Eye Irritation: | 0.264 |
| Respiratory Toxicity: | 0.113 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC002660 | 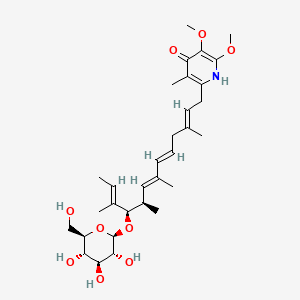 |
0.652 | D0B1IP | 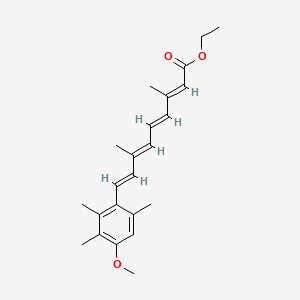 |
0.271 | ||
| ENC003820 | 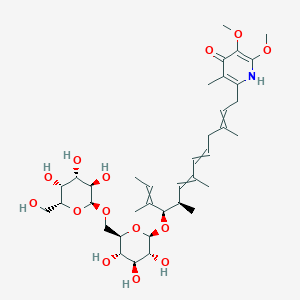 |
0.517 | D05QDC |  |
0.263 | ||
| ENC003819 |  |
0.425 | D0L5FY |  |
0.232 | ||
| ENC004854 |  |
0.375 | D04FBR | 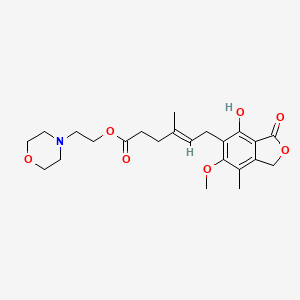 |
0.210 | ||
| ENC004855 | 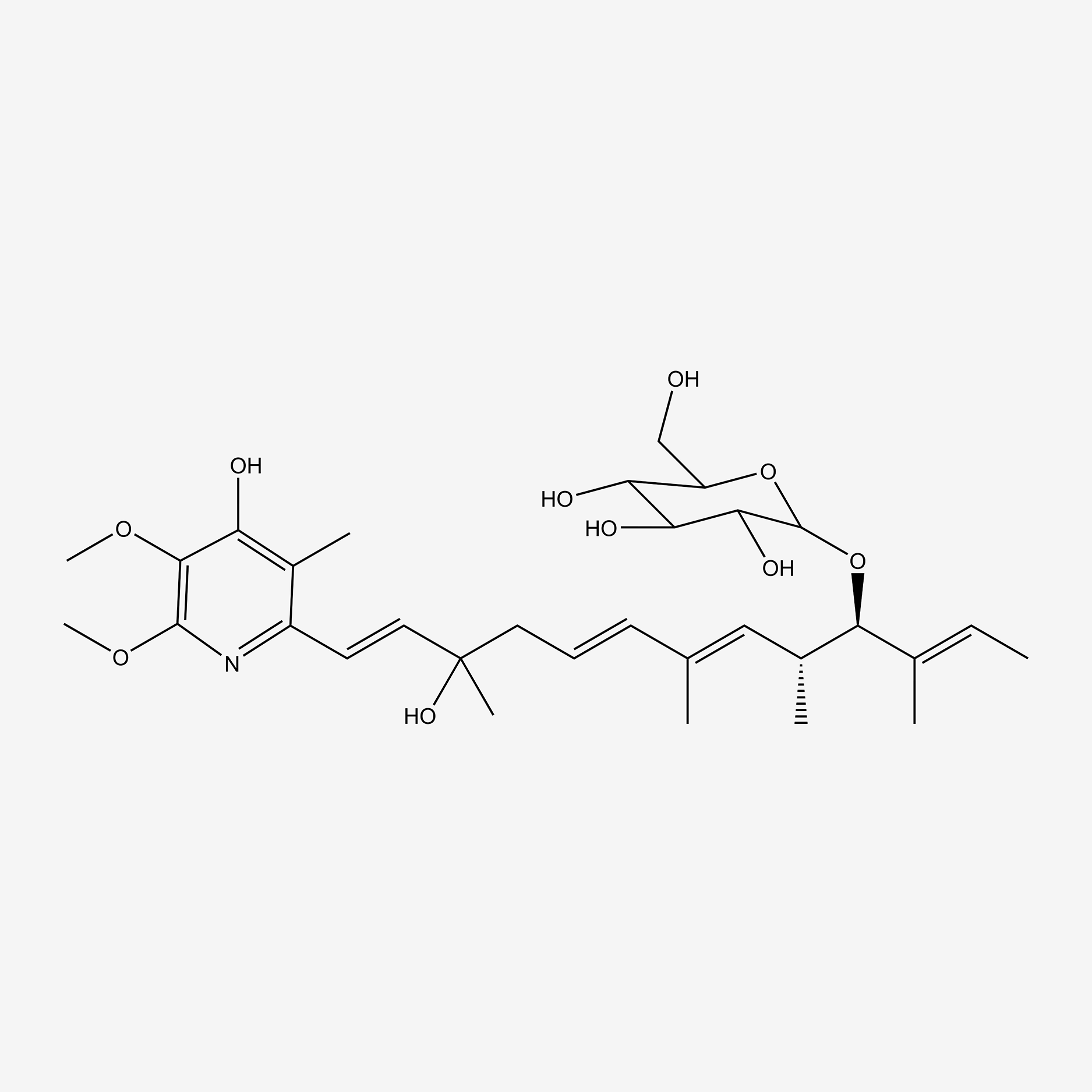 |
0.352 | D0WY9N | 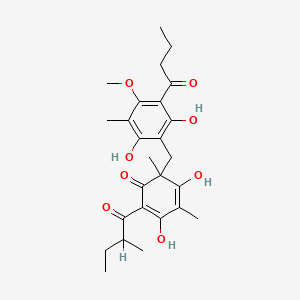 |
0.200 | ||
| ENC005021 | 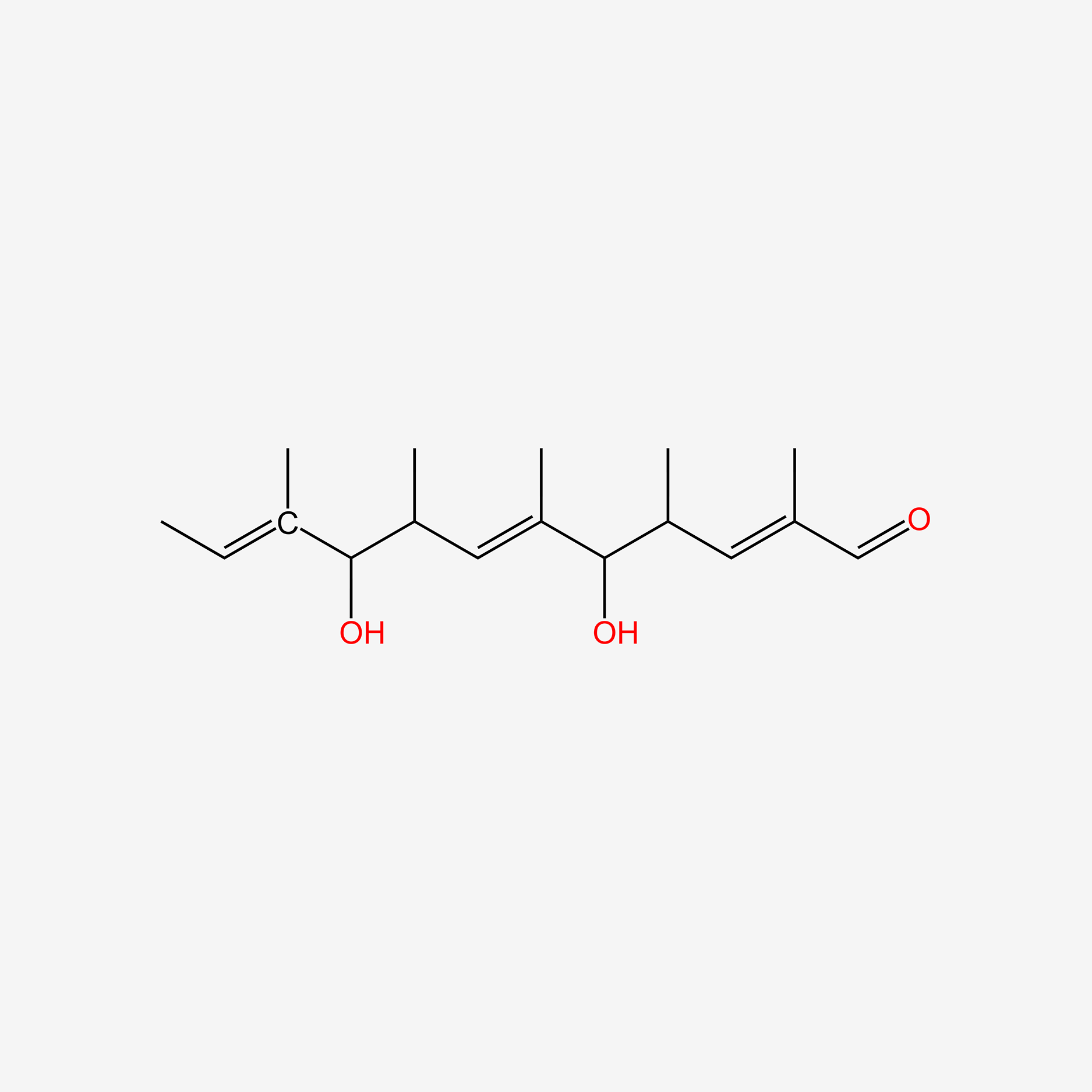 |
0.351 | D0S7WX | 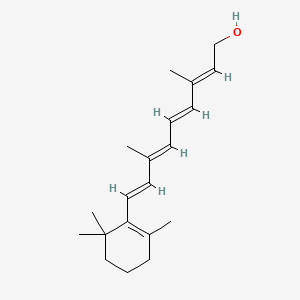 |
0.191 | ||
| ENC004634 |  |
0.309 | D00WVW | 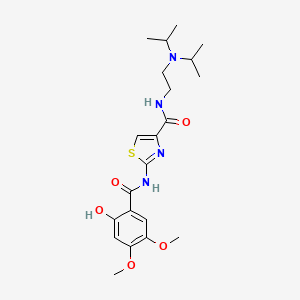 |
0.188 | ||
| ENC006092 |  |
0.291 | D0G3PI | 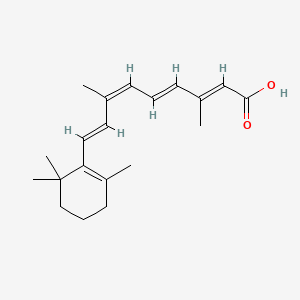 |
0.188 | ||
| ENC005286 |  |
0.288 | D02DGU | 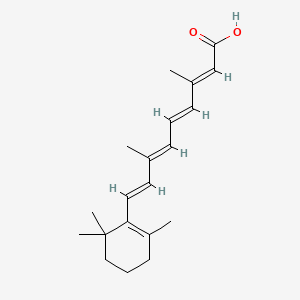 |
0.188 | ||
| ENC005161 |  |
0.272 | D00DKK | 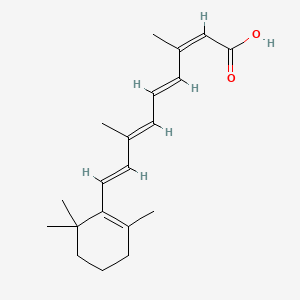 |
0.188 | ||