NPs Basic Information
|
Name |
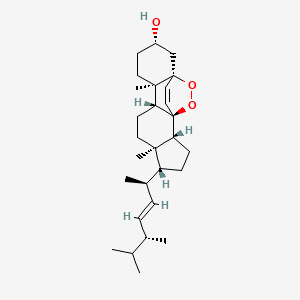
Ergosterol peroxide
|
| Molecular Formula | C28H44O3 | |
| IUPAC Name* |
(1S,2R,5R,6R,9R,10R,13S,15S)-5-[(E,2R,5R)-5,6-dimethylhept-3-en-2-yl]-6,10-dimethyl-16,17-dioxapentacyclo[13.2.2.01,9.02,6.010,15]nonadec-18-en-13-ol
|
|
| SMILES |
C[C@H](/C=C/[C@H](C)C(C)C)[C@H]1CC[C@@H]2[C@@]1(CC[C@H]3[C@]24C=C[C@@]5([C@@]3(CC[C@@H](C5)O)C)OO4)C
|
|
| InChI |
InChI=1S/C28H44O3/c1-18(2)19(3)7-8-20(4)22-9-10-23-25(22,5)13-12-24-26(6)14-11-21(29)17-27(26)15-16-28(23,24)31-30-27/h7-8,15-16,18-24,29H,9-14,17H2,1-6H3/b8-7+/t19-,20+,21-,22+,23+,24+,25+,26+,27+,28-/m0/s1
|
|
| InChIKey |
VXOZCESVZIRHCJ-KGHQQZOUSA-N
|
|
| Synonyms |
Ergosterol peroxide; Ergosterol endoperoxide; Ergosterol-5,8-peroxide; Peroxyergosterol; 2061-64-5; Ergosterol 5alpha,8alpha-epidioxide; UG9TN81TGH; CHEMBL434750; CHEBI:65858; 5alpha,8alpha-epidioxyergosta-6,22-dien-3beta-ol; NSC31324; NSC-31324; (1S,2R,5R,6R,9R,10R,13S,15S)-5-[(E,2R,5R)-5,6-dimethylhept-3-en-2-yl]-6,10-dimethyl-16,17-dioxapentacyclo[13.2.2.01,9.02,6.010,15]nonadec-18-en-13-ol; 5alpha,8alpha-epidioxy-22E-ergosta-6,22-dien-3beta-ol; (22E,24R)-5alpha,8alpha-epidioxyergosta-6,22-dien-3beta-ol; (24R)-5alpha,8alpha-epidioxy-24-methylcholesta-6,22-dien-3beta-ol; NSC 31324; (3S,5S,8S,9R,10R,13R,14R,17R)-17-[(2R,3E,5R)-5,6-dimethylhept-3-en-2-yl]-10,13-dimethyl-1,3,4,9,10,11,12,13,14,15,16,17-dodecahydro-2H-5,8-epidioxycyclopenta[a]phenanthren-3-ol; UNII-UG9TN81TGH; (3beta,5alpha,8alpha,22E)-5,8-epidioxyergosta-6,22-dien-3-ol; 5.alpha.,8.alpha.-Ergosta-6,22-dien-3.beta.-ol, 5,8-epidioxy-; Ergosta-6,22-dien-3-ol, 5,8-epidioxy-, (3.beta.,5.alpha.,8.alpha.,22E)-; Ergosterol 5.alpha.,8.alpha.-epidioxide; Ergosterol 5-alpha,8-alpha-epidioxide; BRN 0096852; ERGOSTERYL PEROXIDE; 5-19-03-00043 (Beilstein Handbook Reference); Ergosterol Peroxide_120246; SCHEMBL3281353; ERGOSTEROL 5,8-PEROXIDE; 5,8-Epidioxy-5-alpha,8-alpha-ergosta-6,22-dien-3-beta-ol; DTXSID501021533; HY-N3845; BDBM50334064; ZINC38139718; ERGOSTEROL ENDOPEROXIDE [INCI]; CS-0024320; 5.ALPHA.,8.ALPHA.-EPIDIOXYERGOSTEROL; 5.alpha.,22-dien-3.beta.-ol, 5,8-epidioxy-; 5alpha,8alpha-Epidioxyergosta-6,22-diene-3beta-ol; Q5385831; 5-alpha,8-alpha-ERGOSTA-6,22-DIEN-3-beta-OL, 5,8-EPIDIOXY-; Ergosta-6, 5,8-epidioxy-, (3.beta.,5.alpha.,8.alpha.,22E)-; (22E)-5.ALPHA.,8.ALPHA.-EPIDIOXYERGOSTA-6,22-DIEN-3.BETA.-OL; (24R)-ERGOST-5.ALPHA.,8.ALPHA.-EPIDIOXY-6,22-DIEN-3.BETA.-OL; 3.BETA.-HYDROXY-5.ALPHA.,8.ALPHA.-EPIDIOXYERGOSTA-6,22-DIENE; Ergosta-6,22-dien-3-ol, 5,8-epidioxy-, (3-beta,5-alpha,8-alpha,22E)-; Ergosta-6,22-dien-3-ol, 5,8-epidioxy-, (3beta,5alpha,8alpha,22E)-; (22E,24R)-5.ALPHA.,8.ALPHA.-EPIDIOXYERGOSTA-6,22-DIEN-3.BETA.-OL; 5.ALPHA.,8.ALPHA.-EPIDOXY-(22E,24R)-ERGOSTA-6,22-DIEN-3.BETA.-OL; (1S,2R,5R,6R,9R,10R,13S,15S)-5-[(2R,3E,5R)-5,6-dimethylhept-3-en-2-yl]-6,10-dimethyl-16,17-dioxapentacyclo[13.2.2.0^{1,9}.0^{2,6}.0^{10,15}]nonadec-18-en-13-ol
|
|
| CAS | 2061-64-5 | |
| PubChem CID | 5351516 | |
| ChEMBL ID | CHEMBL434750 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 428.6 | ALogp: | 6.7 |
| HBD: | 1 | HBA: | 3 |
| Rotatable Bonds: | 4 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 38.7 | Aromatic Rings: | 6 |
| Heavy Atoms: | 31 | QED Weighted: | 0.414 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.849 | MDCK Permeability: | 0.00001820 |
| Pgp-inhibitor: | 0.002 | Pgp-substrate: | 0.663 |
| Human Intestinal Absorption (HIA): | 0.007 | 20% Bioavailability (F20%): | 0.027 |
| 30% Bioavailability (F30%): | 0.038 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.066 | Plasma Protein Binding (PPB): | 90.01% |
| Volume Distribution (VD): | 1.146 | Fu: | 2.06% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.035 | CYP1A2-substrate: | 0.954 |
| CYP2C19-inhibitor: | 0.078 | CYP2C19-substrate: | 0.95 |
| CYP2C9-inhibitor: | 0.265 | CYP2C9-substrate: | 0.034 |
| CYP2D6-inhibitor: | 0.026 | CYP2D6-substrate: | 0.142 |
| CYP3A4-inhibitor: | 0.915 | CYP3A4-substrate: | 0.929 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 3.268 | Half-life (T1/2): | 0.123 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.78 | Human Hepatotoxicity (H-HT): | 0.891 |
| Drug-inuced Liver Injury (DILI): | 0.012 | AMES Toxicity: | 0.003 |
| Rat Oral Acute Toxicity: | 0.778 | Maximum Recommended Daily Dose: | 0.985 |
| Skin Sensitization: | 0.345 | Carcinogencity: | 0.011 |
| Eye Corrosion: | 0.006 | Eye Irritation: | 0.047 |
| Respiratory Toxicity: | 0.983 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC004740 | 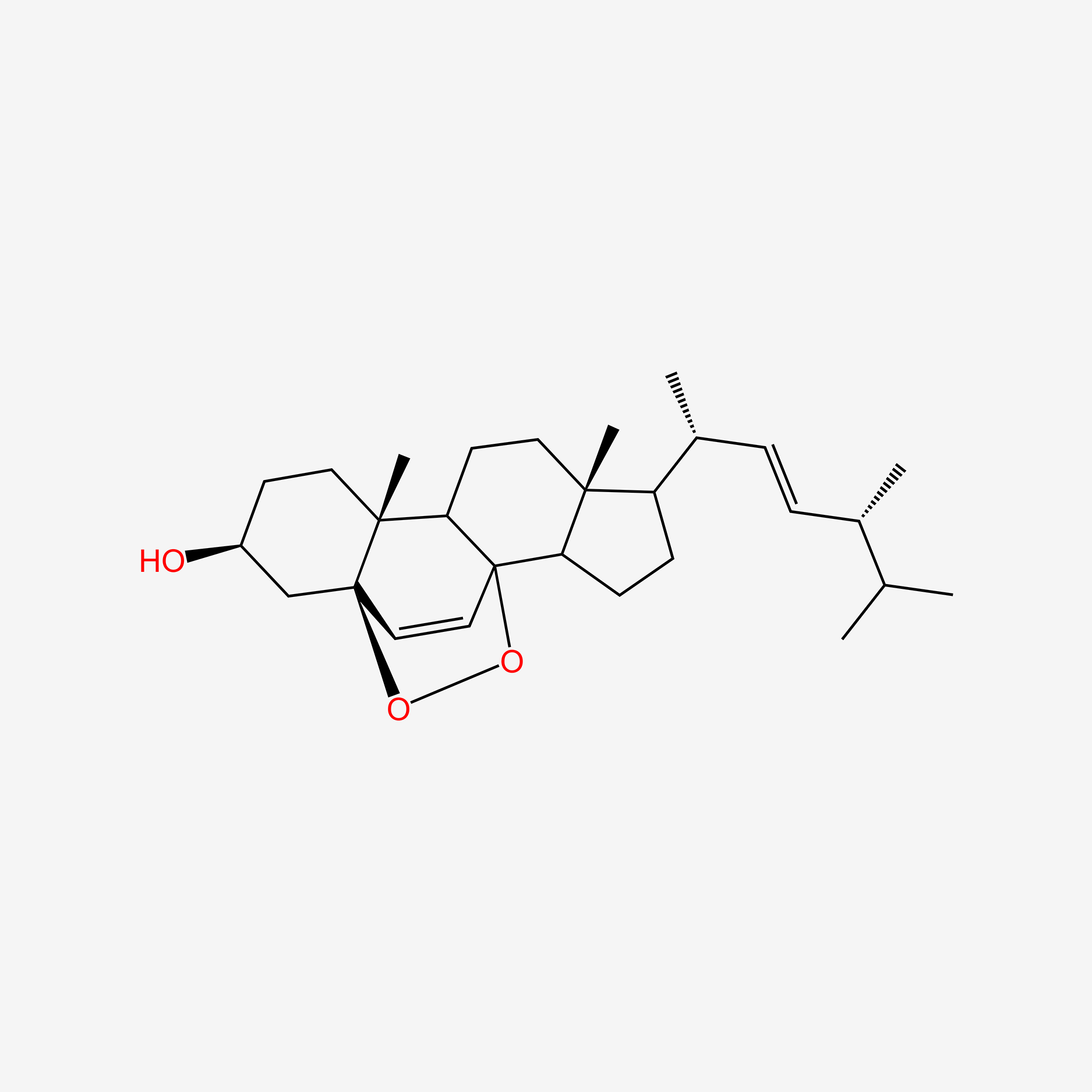 |
1.000 | D0G8OC | 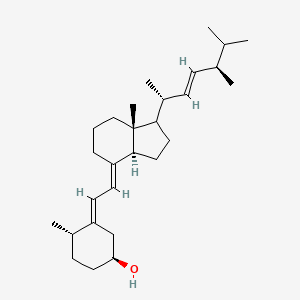 |
0.431 | ||
| ENC005015 | 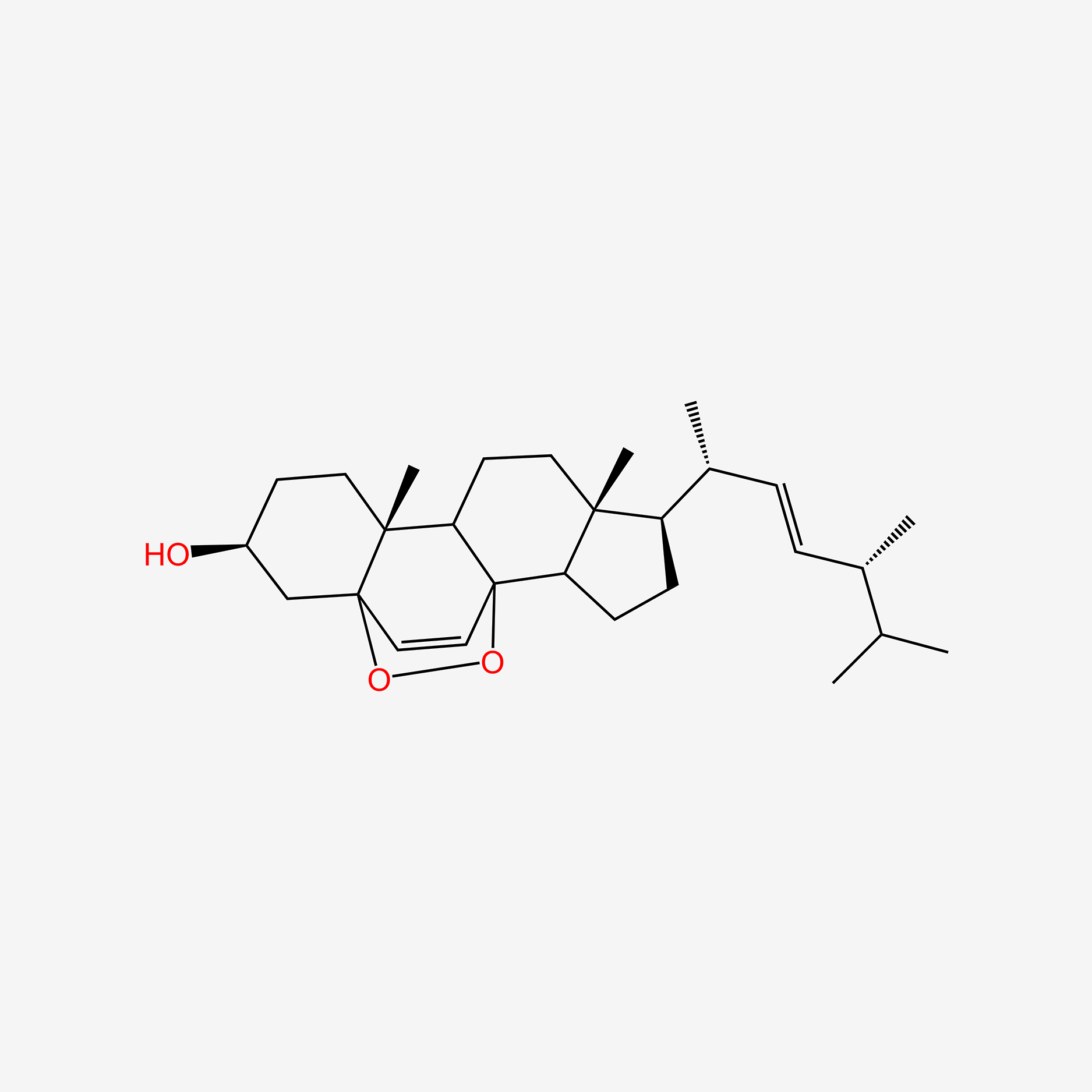 |
1.000 | D06JPB |  |
0.395 | ||
| ENC005013 | 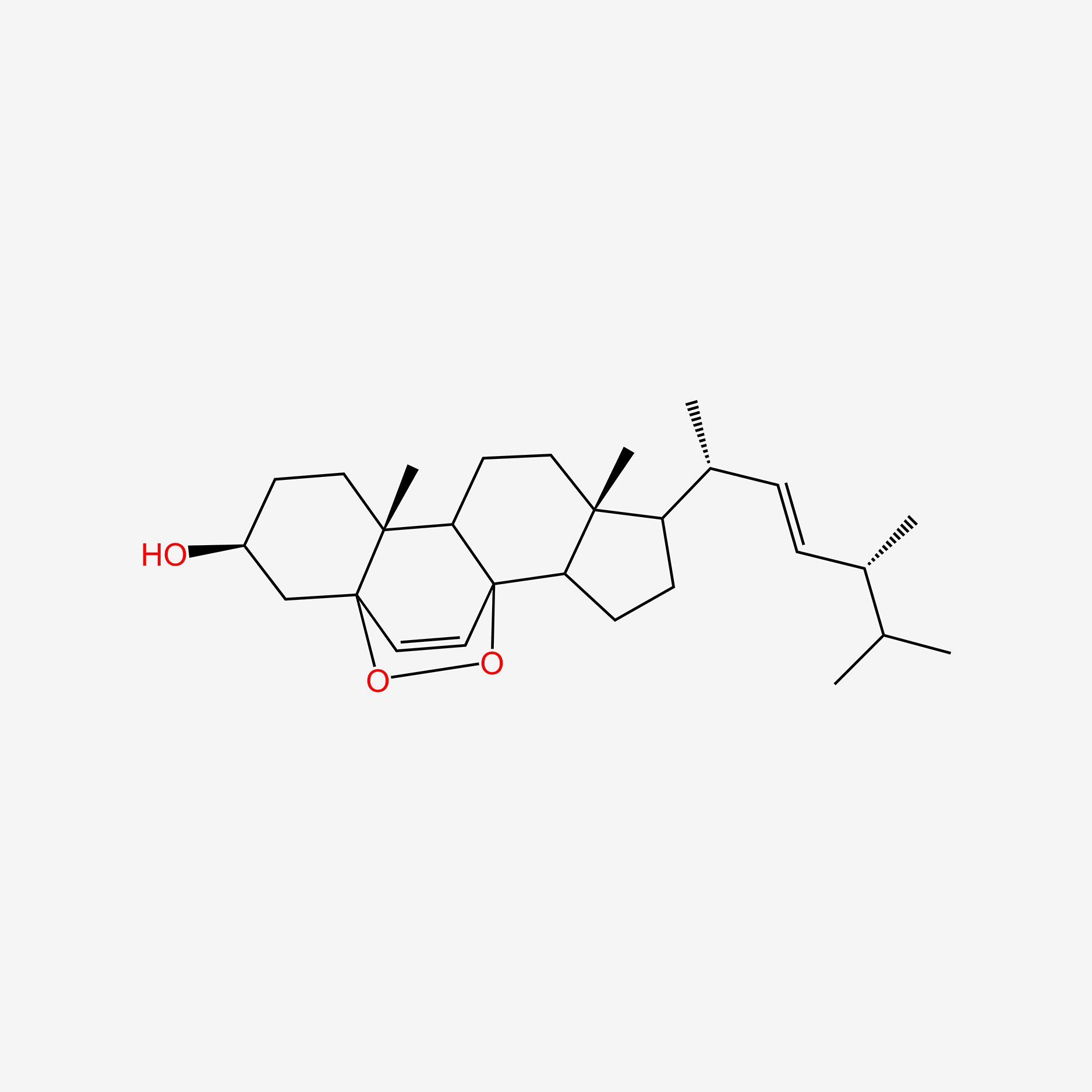 |
1.000 | D0G5CF | 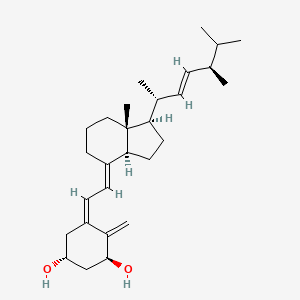 |
0.388 | ||
| ENC005779 |  |
1.000 | D0Y7LD | 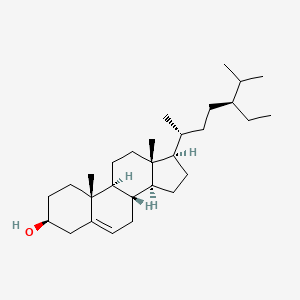 |
0.341 | ||
| ENC004858 | 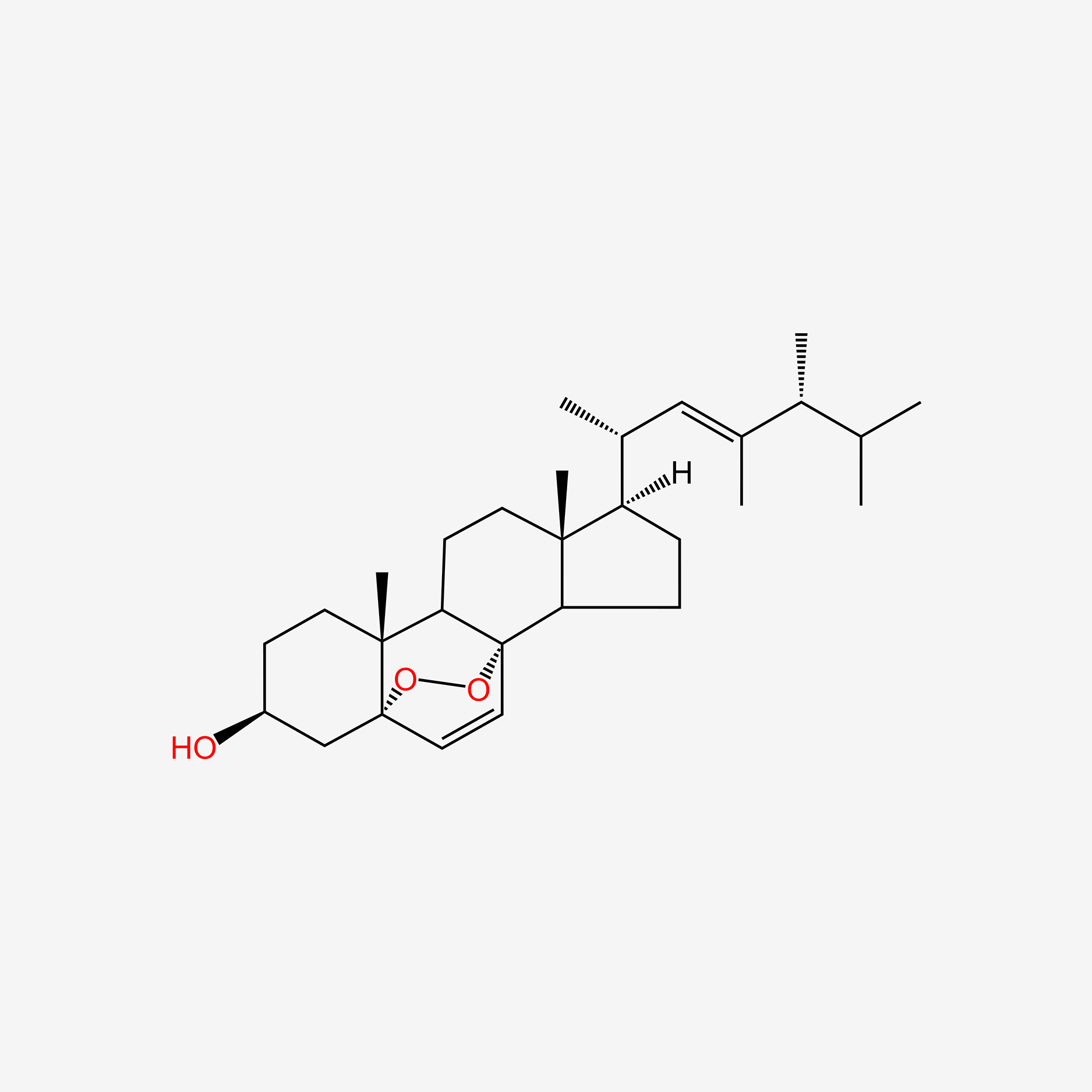 |
0.794 | D0N1TP | 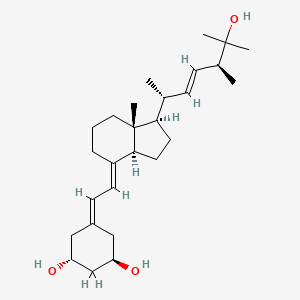 |
0.302 | ||
| ENC002327 | 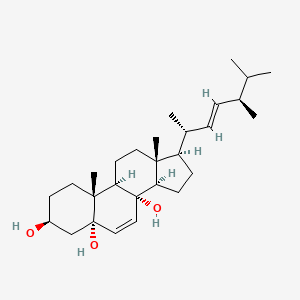 |
0.700 | D01QUS | 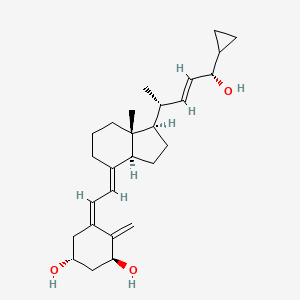 |
0.280 | ||
| ENC004857 | 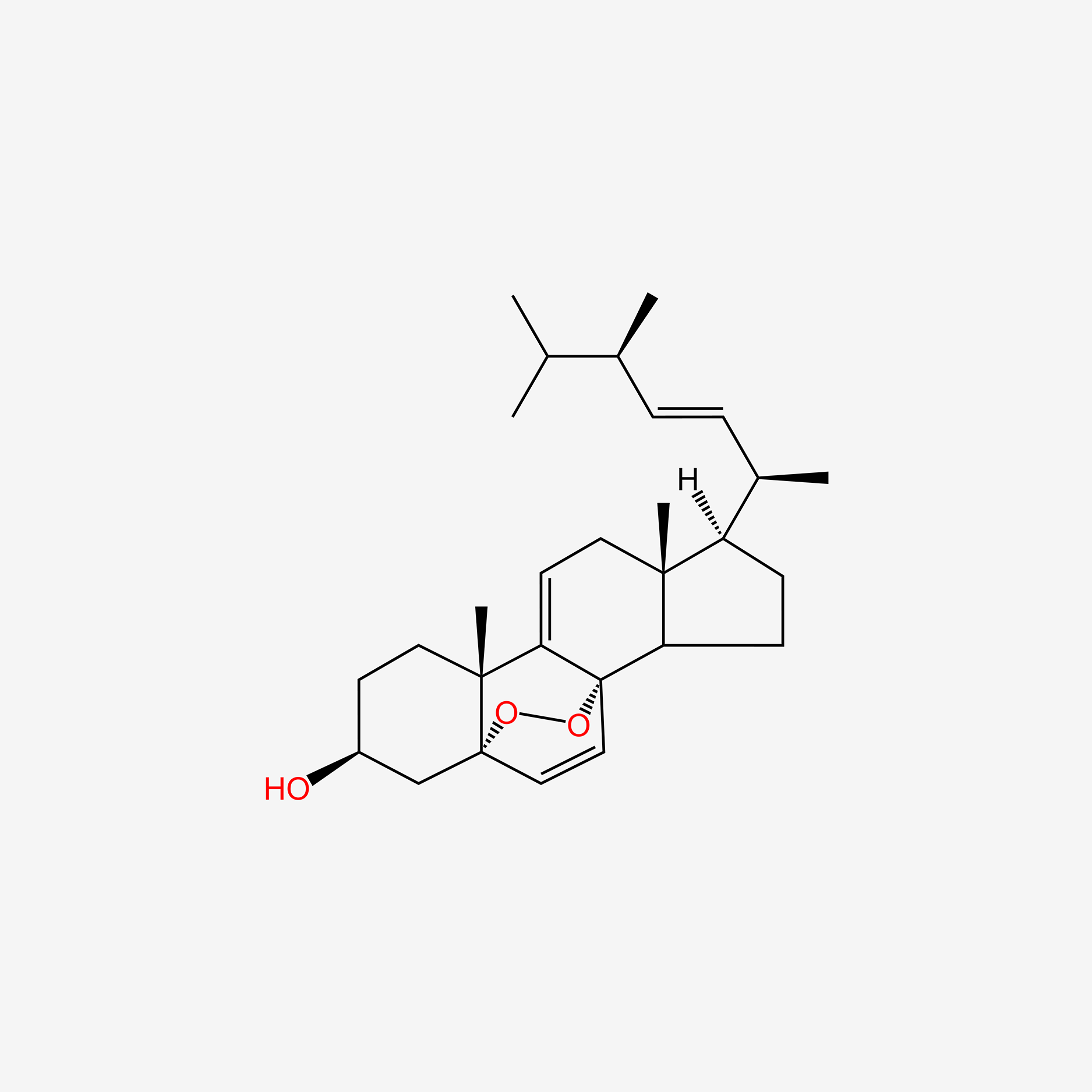 |
0.654 | D0G3SH | 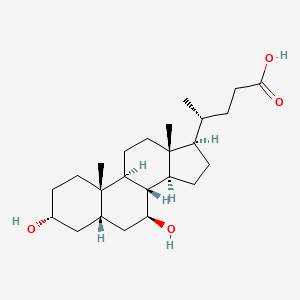 |
0.264 | ||
| ENC005014 | 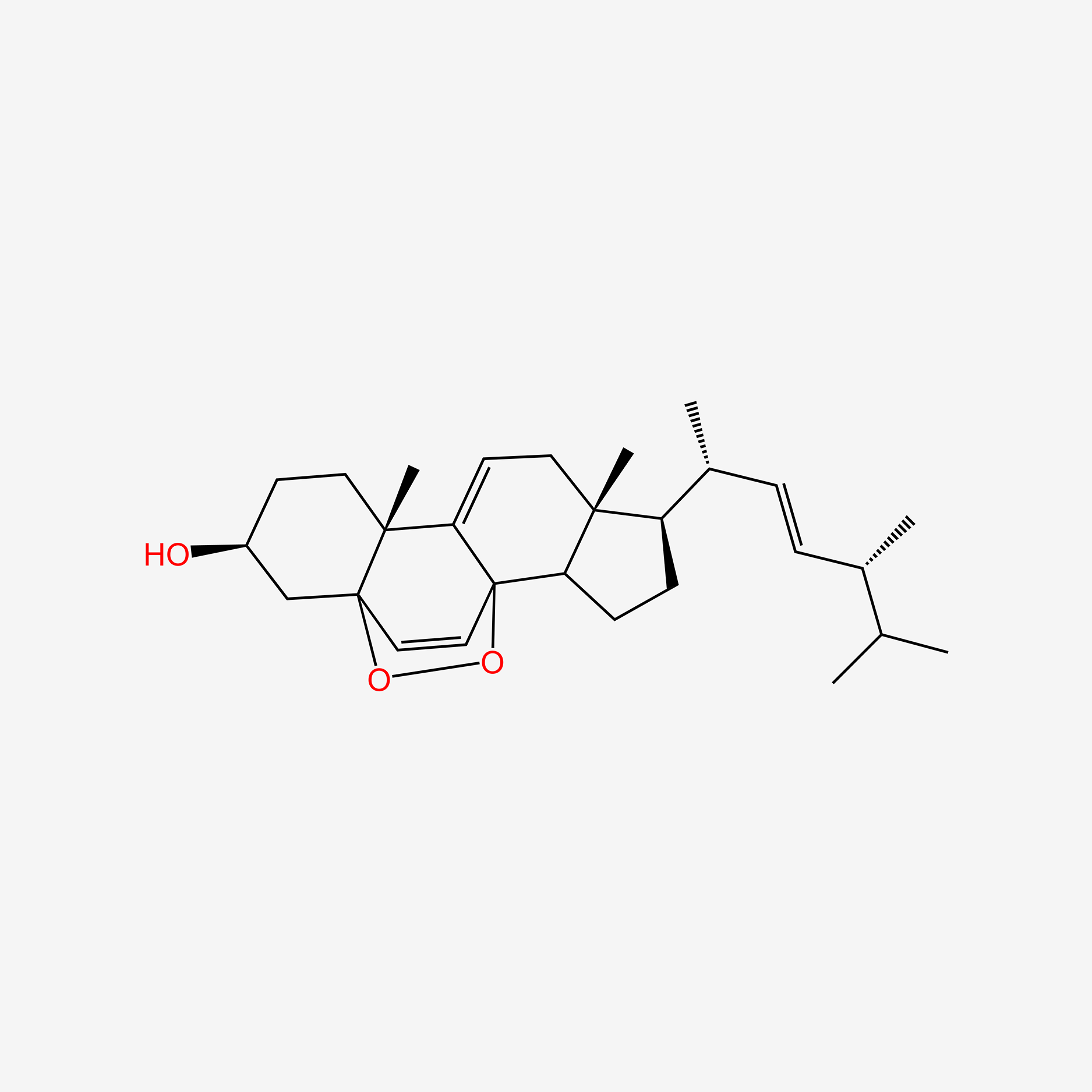 |
0.654 | D03ZTE | 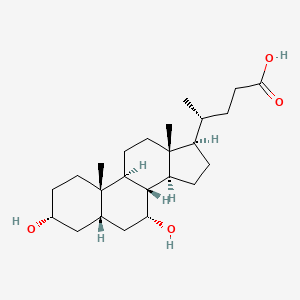 |
0.264 | ||
| ENC004030 | 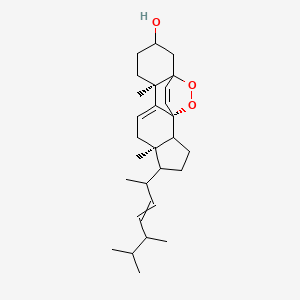 |
0.654 | D0M4WA | 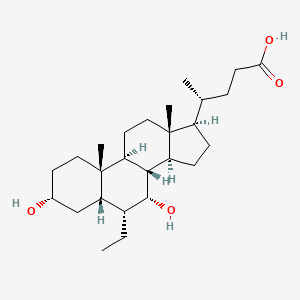 |
0.263 | ||
| ENC006034 | 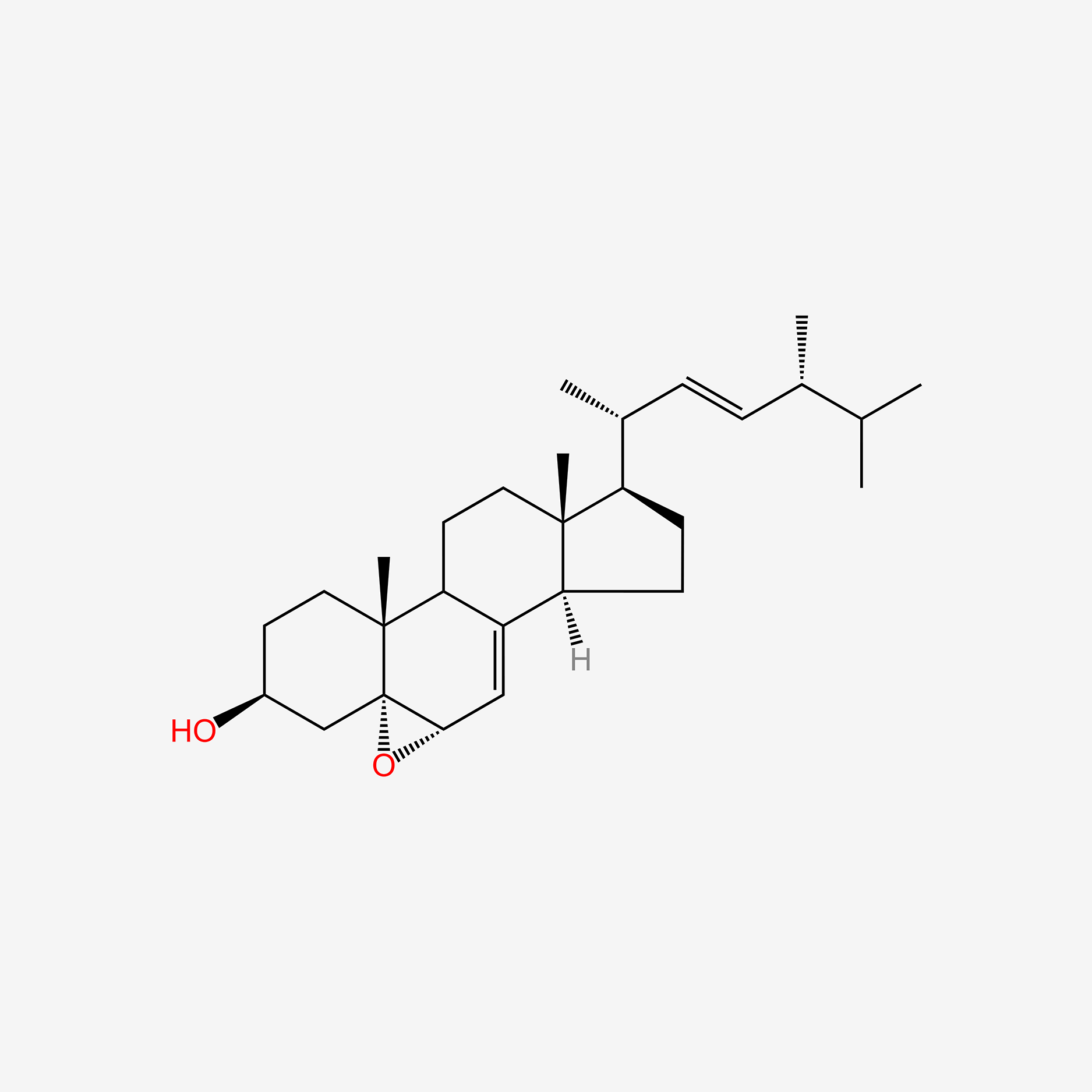 |
0.579 | D08SVH | 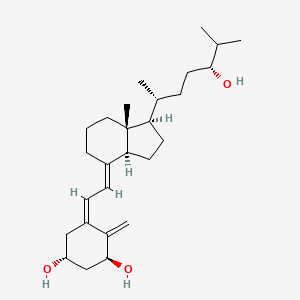 |
0.263 | ||