NPs Basic Information
|
Name |
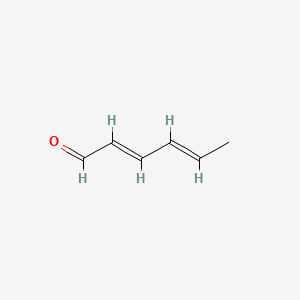
2,4-Hexadienal
|
| Molecular Formula | C6H8O | |
| IUPAC Name* |
(2E,4E)-hexa-2,4-dienal
|
|
| SMILES |
C/C=C/C=C/C=O
|
|
| InChI |
InChI=1S/C6H8O/c1-2-3-4-5-6-7/h2-6H,1H3/b3-2+,5-4+
|
|
| InChIKey |
BATOPAZDIZEVQF-MQQKCMAXSA-N
|
|
| Synonyms |
2,4-HEXADIENAL; 142-83-6; Hexa-2,4-dienal; Sorbaldehyde; Sorbic aldehyde; trans,trans-2,4-Hexadienal; (2E,4E)-hexa-2,4-dienal; 2,4-Hexadienal, (2E,4E)-; (E,E)-2,4-hexadienal; 80466-34-8; (2E,4E)-2,4-Hexadienal; 2,4-Hexadienal, (E,E)-; FEMA No. 3429; trans,trans-Hexa-2,4-dienal; 2,4-Hexadienal, trans,trans-; (E,E)-2,4-Hexadien-1-al; Hexadienal; CHEBI:82334; 878K4I6N7T; 2-Propyleneacrolein; 3-Propyleneacrolein; 2,4-Hexadienal (89% trans,trans-, 11% cis,trans-); 1,3-Pentadiene-1-carboxaldehyde; CCRIS 4030; HSDB 7239; (2E,4E)-Hexadienal; EINECS 205-564-3; 2,4-(e-e)-hexadienal; BRN 1698401; (E,E)-hexa-2,4-dienal; UNII-878K4I6N7T; (E)-2,(E)-4-Hexadienal; AI3-31142; NSC-16184; NSC-68096; trans,trans-2,4-Hexadien-1-al; MFCD00007004; 73506-81-7; e,e-2,4-hexadienal; trans-2,4-hexadienal; t,t-2,4-Hexadienal; (2E,4E) hexadienal; hexadienal (2e,4e-); DSSTox_CID_5391; (2E)-2,4-Hexadienal; 2,4-(E,E)-Hexadienal; DSSTox_RID_77769; DSSTox_GSID_25391; (2e,4z)-2,4-hexadienal; 4-01-00-03545 (Beilstein Handbook Reference); CHEMBL1576086; DTXSID2025391; Hexa-2,4-dienal, (E,E)-; 2,4-HEXADIENAL [HSDB]; (2E,4E)-2,4-Hexadienal #; n-Hex-2,4-dienal, trans,trans-; 2,4-Hexadien-1-al, (E,E)-; trans,trans-2,4-Hexadienal, 95%; ZINC1733897; Tox21_200834; LMFA06000006; AKOS015915466; trans,trans-2,4-Hexadienal, >=95%; NCGC00090786-01; NCGC00090786-02; NCGC00090786-03; NCGC00258388-01; CAS-142-83-6; 2,4-Hexadienal, predominantly trans,trans; DB-003137; TRANS,TRANS-2,4-HEXADIENAL [FHFI]; H0581; C19249; D90909; EN300-113666; trans,trans-2,4-Hexadienal, analytical standard; A807974; A929664; J-007704; Q27155883
|
|
| CAS | 142-83-6 | |
| PubChem CID | 637564 | |
| ChEMBL ID | CHEMBL1576086 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 96.13 | ALogp: | 1.2 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 7 | QED Weighted: | 0.291 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.327 | MDCK Permeability: | 0.00002610 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.01 |
| Human Intestinal Absorption (HIA): | 0.008 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.003 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.996 | Plasma Protein Binding (PPB): | 53.56% |
| Volume Distribution (VD): | 1.301 | Fu: | 49.06% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.207 | CYP1A2-substrate: | 0.439 |
| CYP2C19-inhibitor: | 0.041 | CYP2C19-substrate: | 0.759 |
| CYP2C9-inhibitor: | 0.019 | CYP2C9-substrate: | 0.627 |
| CYP2D6-inhibitor: | 0.008 | CYP2D6-substrate: | 0.876 |
| CYP3A4-inhibitor: | 0.005 | CYP3A4-substrate: | 0.2 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.699 | Half-life (T1/2): | 0.733 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.021 | Human Hepatotoxicity (H-HT): | 0.547 |
| Drug-inuced Liver Injury (DILI): | 0.06 | AMES Toxicity: | 0.805 |
| Rat Oral Acute Toxicity: | 0.968 | Maximum Recommended Daily Dose: | 0.909 |
| Skin Sensitization: | 0.969 | Carcinogencity: | 0.772 |
| Eye Corrosion: | 0.952 | Eye Irritation: | 0.994 |
| Respiratory Toxicity: | 0.955 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001808 |  |
0.429 | D03ZFG |  |
0.145 | ||
| ENC001600 |  |
0.429 | D02DGU |  |
0.114 | ||
| ENC001733 |  |
0.394 | D00DKK |  |
0.114 | ||
| ENC001725 |  |
0.367 | D0G3PI |  |
0.114 | ||
| ENC001724 |  |
0.366 | D0T3NY |  |
0.111 | ||
| ENC001463 |  |
0.353 | D0FG6M |  |
0.111 | ||
| ENC001654 |  |
0.281 | D05QDC | 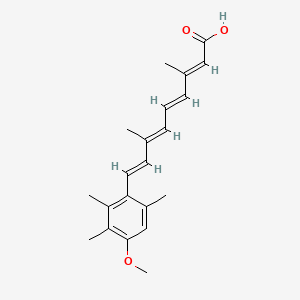 |
0.107 | ||
| ENC003396 | 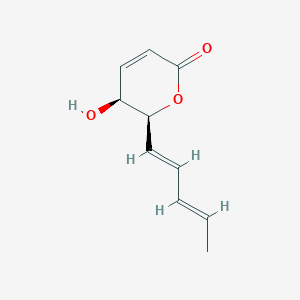 |
0.279 | D0UE9X | 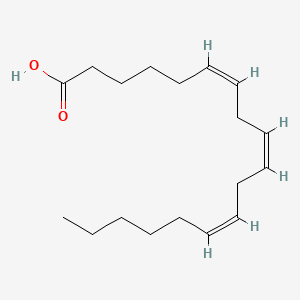 |
0.101 | ||
| ENC001740 |  |
0.263 | D0S7WX | 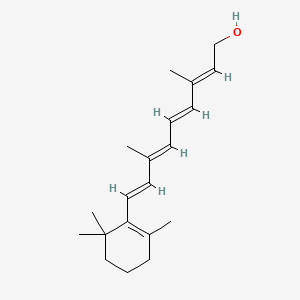 |
0.101 | ||
| ENC001597 | 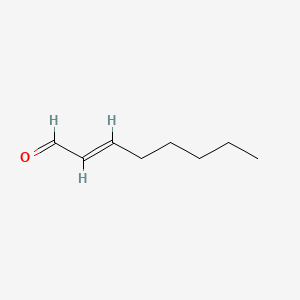 |
0.257 | D0B1IP | 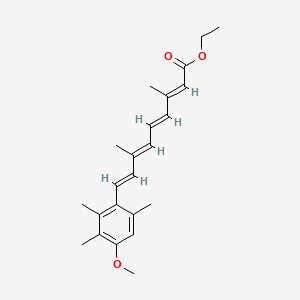 |
0.099 | ||