NPs Basic Information

|
Name |
4-Methyl-6-nitroquinolin-2-ol
|
| Molecular Formula | C10H8N2O3 | |
| IUPAC Name* |
4-methyl-6-nitro-1H-quinolin-2-one
|
|
| SMILES |
CC1=CC(=O)NC2=C1C=C(C=C2)[N+](=O)[O-]
|
|
| InChI |
InChI=1S/C10H8N2O3/c1-6-4-10(13)11-9-3-2-7(12(14)15)5-8(6)9/h2-5H,1H3,(H,11,13)
|
|
| InChIKey |
QBBNQTCDZWMPAH-UHFFFAOYSA-N
|
|
| Synonyms |
4-methyl-6-nitroquinolin-2-ol; 90771-17-8; 4-methyl-6-nitro-1H-quinolin-2-one; 4-Methyl-6-nitro-2(1H)-quinolinone; 2-Hydroxy-4-methyl-6-nitroquinoline; CBMicro_033436; Cambridge id 5796050; Oprea1_082474; Oprea1_318868; SCHEMBL5909911; CHEMBL1886151; DTXSID80346059; ALBB-005438; ZINC9461262; BBL013251; MFCD01014320; STK503410; STK736439; 4-methyl-6-nitroquinolin-2(1H)-one; AKOS000264872; AKOS003388296; SB68740; NCGC00188260-01; 4-Methyl-6-nitro-2(1H)-quinolinone #; VS-03721; BIM-0033578.P001; BB 0260320; CS-0257735; 1,2-dihydro-4-methyl-6-nitro-2-oxo quinoline; 1,2-Dihydro-4-methyl-6-nitro-2-oxoquinoline; D84402; EN300-6763387; 4-METHYL-6-NITRO-1,2-DIHYDROQUINOLIN-2-ONE; Z57464802
|
|
| CAS | 90771-17-8 | |
| PubChem CID | 609262 | |
| ChEMBL ID | CHEMBL1886151 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 204.18 | ALogp: | 1.1 |
| HBD: | 1 | HBA: | 3 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 74.9 | Aromatic Rings: | 2 |
| Heavy Atoms: | 15 | QED Weighted: | 0.572 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.646 | MDCK Permeability: | 0.00009520 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.006 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.001 |
| 30% Bioavailability (F30%): | 0.001 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.404 | Plasma Protein Binding (PPB): | 88.69% |
| Volume Distribution (VD): | 0.607 | Fu: | 6.24% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.967 | CYP1A2-substrate: | 0.865 |
| CYP2C19-inhibitor: | 0.311 | CYP2C19-substrate: | 0.084 |
| CYP2C9-inhibitor: | 0.057 | CYP2C9-substrate: | 0.706 |
| CYP2D6-inhibitor: | 0.058 | CYP2D6-substrate: | 0.482 |
| CYP3A4-inhibitor: | 0.066 | CYP3A4-substrate: | 0.146 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.385 | Half-life (T1/2): | 0.431 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.04 | Human Hepatotoxicity (H-HT): | 0.573 |
| Drug-inuced Liver Injury (DILI): | 0.906 | AMES Toxicity: | 0.981 |
| Rat Oral Acute Toxicity: | 0.177 | Maximum Recommended Daily Dose: | 0.728 |
| Skin Sensitization: | 0.92 | Carcinogencity: | 0.885 |
| Eye Corrosion: | 0.062 | Eye Irritation: | 0.968 |
| Respiratory Toxicity: | 0.95 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000798 |  |
0.351 | D0T5WK |  |
0.329 | ||
| ENC001539 |  |
0.351 | D0J9ZR |  |
0.324 | ||
| ENC000034 |  |
0.340 | D0CP4E |  |
0.321 | ||
| ENC001364 |  |
0.315 | D0W2NM |  |
0.313 | ||
| ENC004685 |  |
0.288 | D0Y7PG |  |
0.301 | ||
| ENC000118 | 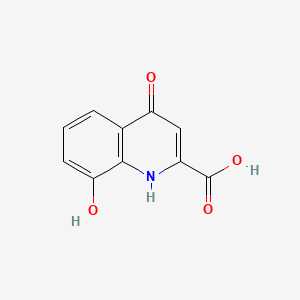 |
0.281 | D0A1DH |  |
0.291 | ||
| ENC002926 | 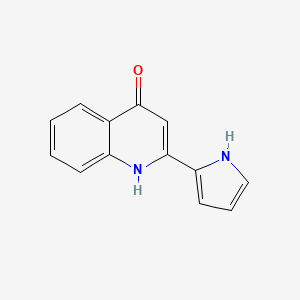 |
0.275 | D0IT2X |  |
0.288 | ||
| ENC005178 | 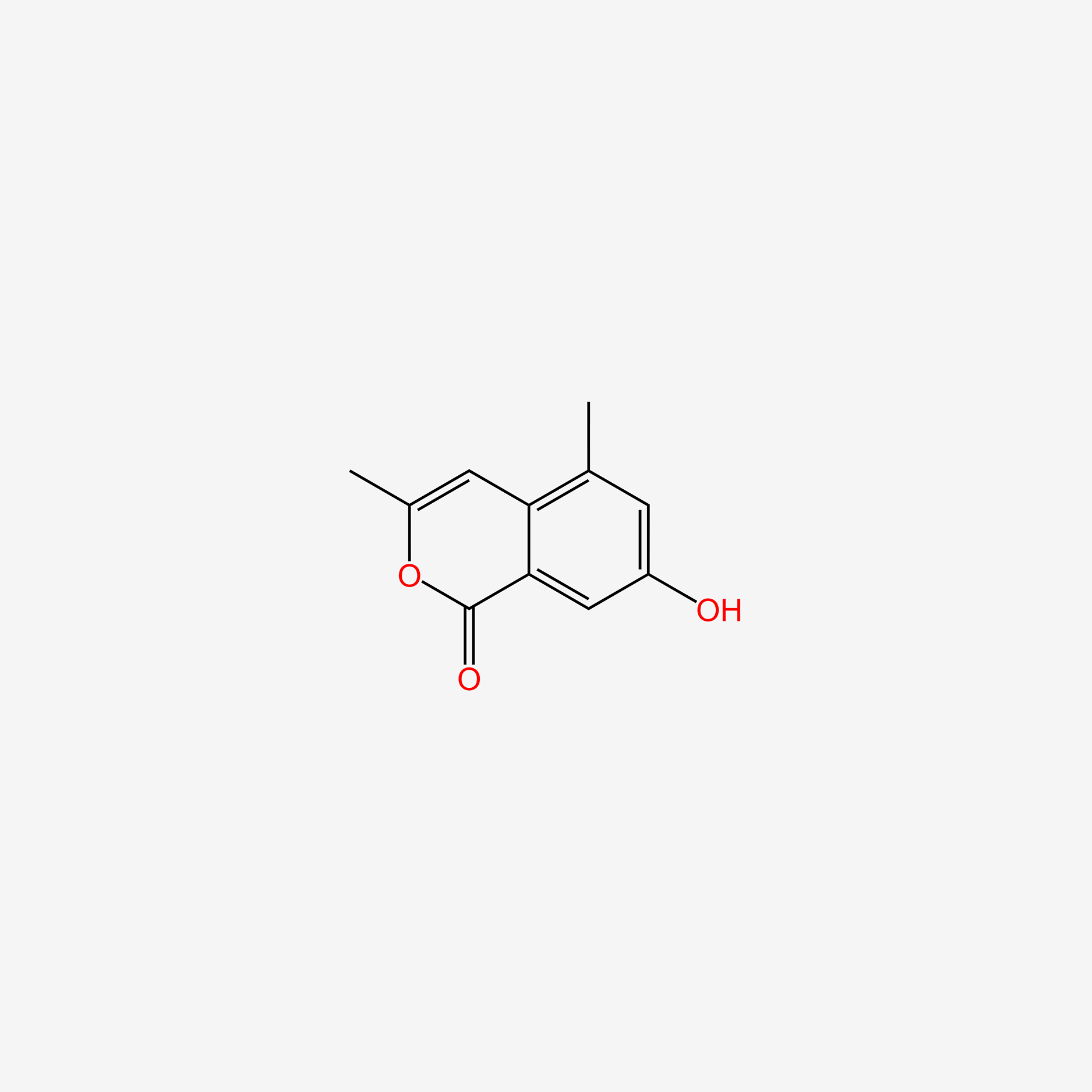 |
0.274 | D0I8DD |  |
0.277 | ||
| ENC000178 | 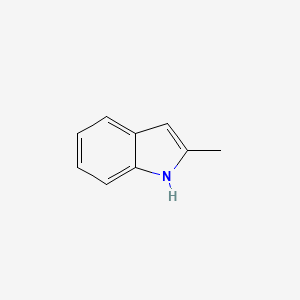 |
0.273 | D0SN9T |  |
0.273 | ||
| ENC005445 |  |
0.273 | D05HFY |  |
0.270 | ||