NPs Basic Information

|
Name |
p-Mentha-1,3,8-triene
|
| Molecular Formula | C10H14 | |
| IUPAC Name* |
1-methyl-4-prop-1-en-2-ylcyclohexa-1,3-diene
|
|
| SMILES |
CC1=CC=C(CC1)C(=C)C
|
|
| InChI |
InChI=1S/C10H14/c1-8(2)10-6-4-9(3)5-7-10/h4,6H,1,5,7H2,2-3H3
|
|
| InChIKey |
XNMPFDIYAMOYRM-UHFFFAOYSA-N
|
|
| Synonyms |
p-Mentha-1,3,8-triene; 1,3,8-p-Menthatriene; p-Menthatriene; 18368-95-1; 1-methyl-4-prop-1-en-2-ylcyclohexa-1,3-diene; p-1,3,8-Menthatriene; p-Menta-1,3,8-triene; 1,3,8-para-Menthatriene; 1,3,8-menthatriene; 1,3-Cyclohexadiene, 1-methyl-4-(1-methylethenyl)-; ZDY5H4QWH3; 1-methyl-4-(prop-1-en-2-yl)cyclohexa-1,3-diene; 1-Isopropenyl-4-methyl-1,3-cyclohexadiene; P-MENTHATRIENE,P-MENTHA-1,3,8-TRIENE; 2-Methyl-5-(1-methylethenyl)-1,3-Cyclohexadiene; UNII-ZDY5H4QWH3; CHEBI:89242; DTXSID60171462; ZINC59585784; 1-Isopropenyl-4-methyl-1,3-cyclohexadiene #; 1-Methyl-4-(1-methylethenyl)-1,3-cyclohexadiene; Q27161428; 1-Methyl-4-(1-methylethenyl)-1,3-cyclohexadiene, 9CI
|
|
| CAS | 18368-95-1 | |
| PubChem CID | 176983 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 134.22 | ALogp: | 3.0 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.508 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.434 | MDCK Permeability: | 0.00001280 |
| Pgp-inhibitor: | 0.009 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.002 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.001 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.366 | Plasma Protein Binding (PPB): | 84.72% |
| Volume Distribution (VD): | 2.517 | Fu: | 23.74% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.783 | CYP1A2-substrate: | 0.909 |
| CYP2C19-inhibitor: | 0.356 | CYP2C19-substrate: | 0.893 |
| CYP2C9-inhibitor: | 0.356 | CYP2C9-substrate: | 0.352 |
| CYP2D6-inhibitor: | 0.345 | CYP2D6-substrate: | 0.824 |
| CYP3A4-inhibitor: | 0.01 | CYP3A4-substrate: | 0.376 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 1.271 | Half-life (T1/2): | 0.713 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.031 | Human Hepatotoxicity (H-HT): | 0.633 |
| Drug-inuced Liver Injury (DILI): | 0.029 | AMES Toxicity: | 0.022 |
| Rat Oral Acute Toxicity: | 0.061 | Maximum Recommended Daily Dose: | 0.238 |
| Skin Sensitization: | 0.949 | Carcinogencity: | 0.694 |
| Eye Corrosion: | 0.933 | Eye Irritation: | 0.983 |
| Respiratory Toxicity: | 0.578 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000198 |  |
0.459 | D0O1UZ | 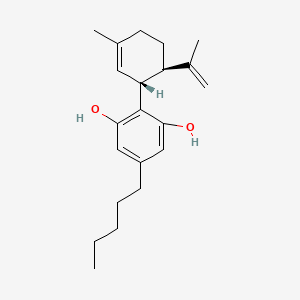 |
0.200 | ||
| ENC002218 | 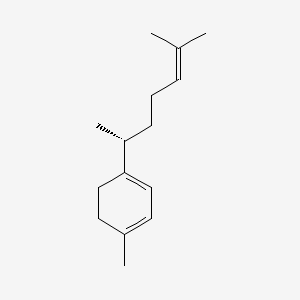 |
0.388 | D03KEK | 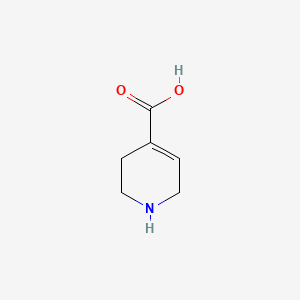 |
0.156 | ||
| ENC002403 |  |
0.350 | D0N0OU | 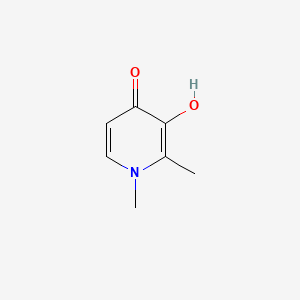 |
0.152 | ||
| ENC000395 |  |
0.350 | D0WO8W |  |
0.149 | ||
| ENC002138 |  |
0.333 | D06XWB |  |
0.149 | ||
| ENC001066 |  |
0.317 | D0Z8SF |  |
0.149 | ||
| ENC000555 |  |
0.317 | D0X0WU |  |
0.147 | ||
| ENC002219 |  |
0.286 | D0H6VY |  |
0.145 | ||
| ENC000194 |  |
0.273 | D04GJN |  |
0.145 | ||
| ENC001484 | 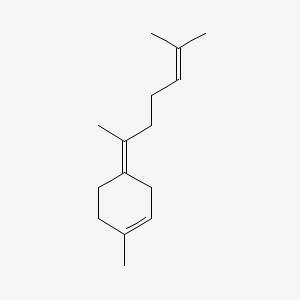 |
0.259 | D01PJR | 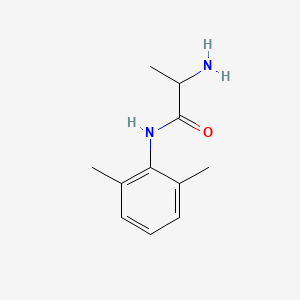 |
0.143 | ||