NPs Basic Information
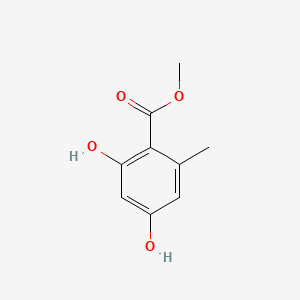
|
Name |
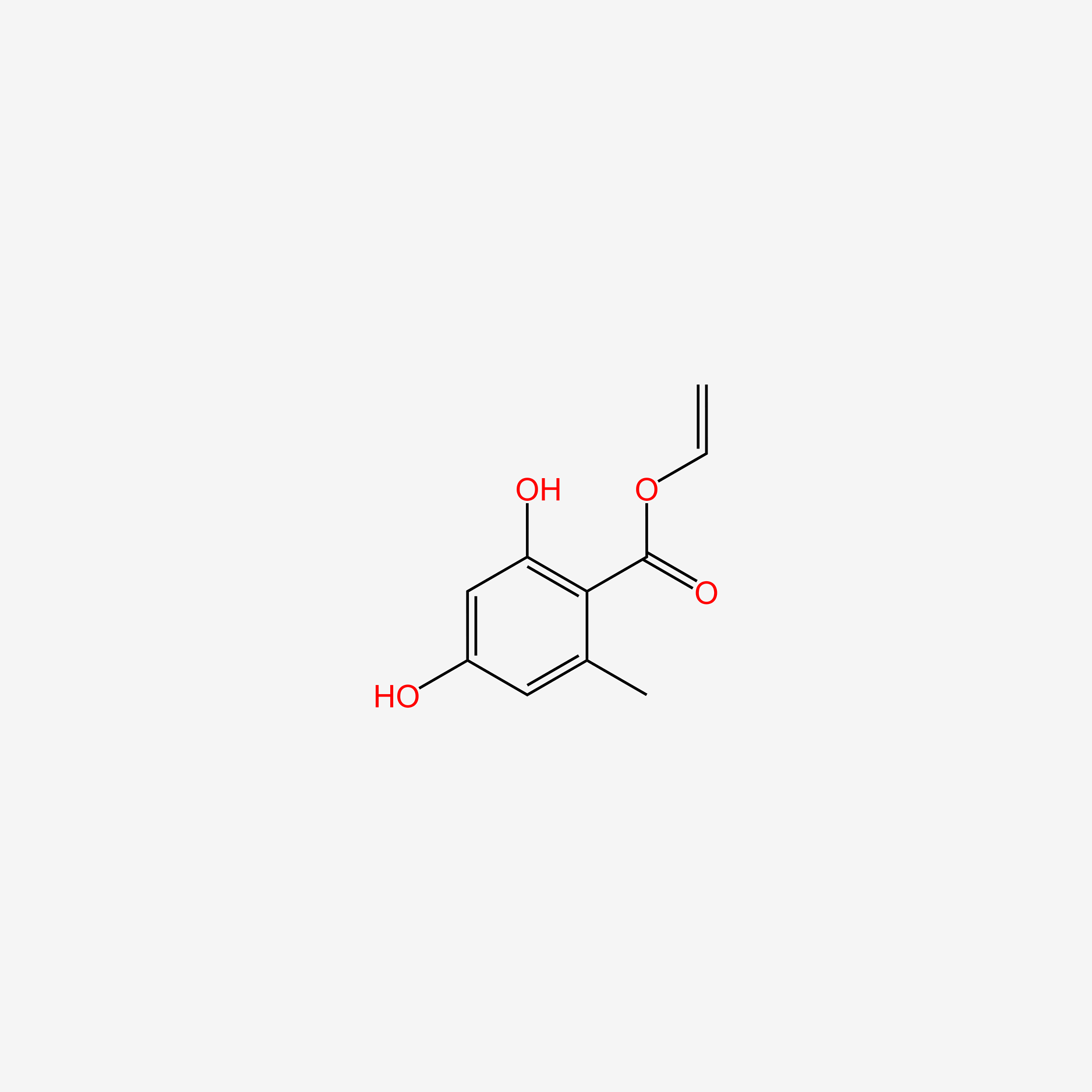
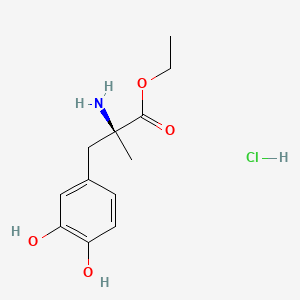
Methyl 2,4-dihydroxy-6-methylbenzoate
|
| Molecular Formula | C9H10O4 | |
| IUPAC Name* |
methyl 2,4-dihydroxy-6-methylbenzoate
|
|
| SMILES |
CC1=CC(=CC(=C1C(=O)OC)O)O
|
|
| InChI |
InChI=1S/C9H10O4/c1-5-3-6(10)4-7(11)8(5)9(12)13-2/h3-4,10-11H,1-2H3
|
|
| InChIKey |
NCCWCZLEACWJIN-UHFFFAOYSA-N
|
|
| Synonyms |
Methyl orsellinate; Methyl 2,4-dihydroxy-6-methylbenzoate; 3187-58-4; methylorsellinate; Benzoic acid, 2,4-dihydroxy-6-methyl-, methyl ester; orsellinic acid methyl ester; Methyl 2,4-dihydroxy-6-methyl benzoate; CHEMBL454431; Orsellinic acid monomethyl ester; CHEBI:67898; 2,4-Dihydroxy-6-methylbenzoic acid methyl ester; Methyl 4,6-dihydroxy-2-methylbenzoate; Methyl o-orsellinate; Orsellinic acid methyl; Spectrum2_001751; .beta.-Resorcylic acid, 6-methyl-, methyl ester; SPECTRUM210925; MLS000728678; SCHEMBL370816; Benzoic acid,2,4-dihydroxy-6-methyl-, methyl ester; SPBio_001882; Methyl 6-methyl-ss-resorcylate; DTXSID2062901; HMS2267N11; ZINC391406; BDBM50294528; CCG-39451; AKOS000349113; methyl 2,4-dihydroxy-6-methyl-benzoate; NCGC00095797-01; BS-25248; SMR000470886; BB 0218022; CS-0023752; SR-01000761473; SR-01000761473-2; BRD-K12202814-001-05-8; Q27136372
|
|
| CAS | 3187-58-4 | |
| PubChem CID | 76658 | |
| ChEMBL ID | CHEMBL454431 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Physi-Chem Properties
| Molecular Weight: | 182.17 | ALogp: | 2.0 |
| HBD: | 2 | HBA: | 4 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 66.8 | Aromatic Rings: | 1 |
| Heavy Atoms: | 13 | QED Weighted: | 0.648 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.649 | MDCK Permeability: | 0.00001380 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.029 |
| Human Intestinal Absorption (HIA): | 0.011 | 20% Bioavailability (F20%): | 0.148 |
| 30% Bioavailability (F30%): | 0.197 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.249 | Plasma Protein Binding (PPB): | 88.49% |
| Volume Distribution (VD): | 0.797 | Fu: | 13.92% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.972 | CYP1A2-substrate: | 0.868 |
| CYP2C19-inhibitor: | 0.233 | CYP2C19-substrate: | 0.086 |
| CYP2C9-inhibitor: | 0.192 | CYP2C9-substrate: | 0.894 |
| CYP2D6-inhibitor: | 0.491 | CYP2D6-substrate: | 0.514 |
| CYP3A4-inhibitor: | 0.625 | CYP3A4-substrate: | 0.177 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 14.547 | Half-life (T1/2): | 0.886 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.023 | Human Hepatotoxicity (H-HT): | 0.054 |
| Drug-inuced Liver Injury (DILI): | 0.513 | AMES Toxicity: | 0.148 |
| Rat Oral Acute Toxicity: | 0.029 | Maximum Recommended Daily Dose: | 0.524 |
| Skin Sensitization: | 0.548 | Carcinogencity: | 0.032 |
| Eye Corrosion: | 0.275 | Eye Irritation: | 0.98 |
| Respiratory Toxicity: | 0.501 |