NPs Basic Information
|
Name |
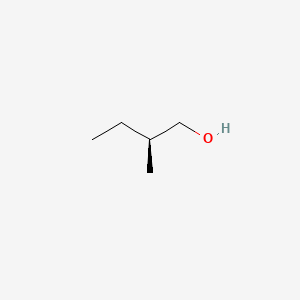
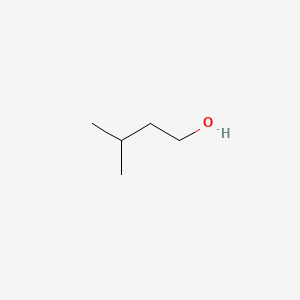
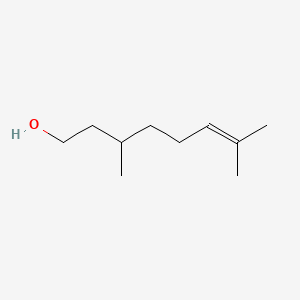
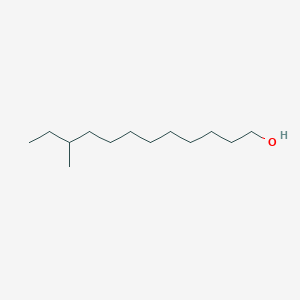
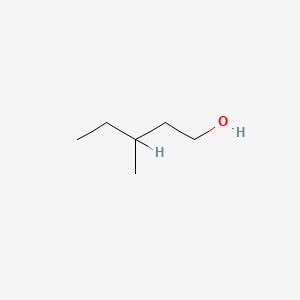
3-Methyl-1-pentanol
|
| Molecular Formula | C6H14O | |
| IUPAC Name* |
3-methylpentan-1-ol
|
|
| SMILES |
CCC(C)CCO
|
|
| InChI |
InChI=1S/C6H14O/c1-3-6(2)4-5-7/h6-7H,3-5H2,1-2H3
|
|
| InChIKey |
IWTBVKIGCDZRPL-UHFFFAOYSA-N
|
|
| Synonyms |
3-METHYL-1-PENTANOL; 3-Methylpentan-1-ol; 589-35-5; 3-Methylpentanol; 2-Ethyl-4-butanol; 3-Ethyl-1-butanol; 1-Pentanol, 3-methyl-; FEMA No. 3762; N8W93SI0FS; (+/-)-3-methyl-1-pentanol; NSC-9466; 20281-83-8; UNII-N8W93SI0FS; NSC9466; 3-methyl pentanol; NSC 9466; EINECS 209-644-9; MFCD00002937; 3-methyl pentan-1-ol; 3-Methyl-pentan-1-ol; AI3-38563; SCHEMBL14990; 3-Methyl-1-pentanol, 99%; SCHEMBL8988208; CHEBI:87381; DTXSID20862248; 3-Methyl-(.+/-.)-1-Pentanol; (DL)-3-METHYLPENTYL ALCOHOL; LMFA05000111; 3-Methyl-1-pentanol, >=99%, FG; AKOS009156537; 3-METHYL-1-PENTANOL, 97%,; SB83830; 1-Pentanol, 3-methyl-, (.+/-.)-; BS-23125; DB-050838; CS-0204814; FT-0616038; FT-0639759; M0600; D91364; EN300-140256; A869318; Q3278324; 343268-11-1
|
|
| CAS | 589-35-5 | |
| PubChem CID | 11508 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Physi-Chem Properties
| Molecular Weight: | 102.17 | ALogp: | 1.8 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 20.2 | Aromatic Rings: | 0 |
| Heavy Atoms: | 7 | QED Weighted: | 0.577 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.143 | MDCK Permeability: | 0.00002810 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.063 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.007 |
| 30% Bioavailability (F30%): | 0.015 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.981 | Plasma Protein Binding (PPB): | 33.81% |
| Volume Distribution (VD): | 1.176 | Fu: | 60.40% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.382 | CYP1A2-substrate: | 0.771 |
| CYP2C19-inhibitor: | 0.045 | CYP2C19-substrate: | 0.649 |
| CYP2C9-inhibitor: | 0.03 | CYP2C9-substrate: | 0.358 |
| CYP2D6-inhibitor: | 0.006 | CYP2D6-substrate: | 0.134 |
| CYP3A4-inhibitor: | 0.011 | CYP3A4-substrate: | 0.203 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.376 | Half-life (T1/2): | 0.775 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.019 | Human Hepatotoxicity (H-HT): | 0.03 |
| Drug-inuced Liver Injury (DILI): | 0.036 | AMES Toxicity: | 0.007 |
| Rat Oral Acute Toxicity: | 0.034 | Maximum Recommended Daily Dose: | 0.036 |
| Skin Sensitization: | 0.393 | Carcinogencity: | 0.254 |
| Eye Corrosion: | 0.984 | Eye Irritation: | 0.992 |
| Respiratory Toxicity: | 0.092 |