NPs Basic Information

|
Name |
Isovaleramide
|
| Molecular Formula | C5H11NO | |
| IUPAC Name* |
3-methylbutanamide
|
|
| SMILES |
CC(C)CC(=O)N
|
|
| InChI |
InChI=1S/C5H11NO/c1-4(2)3-5(6)7/h4H,3H2,1-2H3,(H2,6,7)
|
|
| InChIKey |
SANOUVWGPVYVAV-UHFFFAOYSA-N
|
|
| Synonyms |
ISOVALERAMIDE; 3-Methylbutanamide; 541-46-8; Butanamide, 3-methyl-; 3-Methylbutyramide; Isopentanamide; Isovaleric amide; Isovaleric acid amide; Isovaleramide [USAN]; NFS1776; .beta.-Methylbutyramide; NSC-402555; 9CP4KB634M; NFS-1776; NPS-1776; Isovaleramide (USAN); beta-Methylbutyramide; NPS 1776; EINECS 208-781-1; NSC 402555; BRN 1740789; UNII-9CP4KB634M; 3-methylbutaneamide; ISOVALERAMIDE [MI]; SCHEMBL9641; CHEMBL171066; DTXSID3060249; HMS3264B03; HMS3652O16; HMS3885H05; Pharmakon1600-01506177; ZINC158116; TRIMETHYLENEDI(THIOTOSYLATE); BCP05941; HY-B1229; BDBM50224817; MFCD00014807; NSC402555; NSC760408; s4116; AKOS008937702; CCG-213627; CS-4876; NSC-760408; AS-14001; FT-0627531; SW220069-1; C75541; D04637; AB01563345_01; AB01563345_02; EN300-1232898; 541I468; A870532; J-512895; Q6086595; Z33546511; F1995-0350
|
|
| CAS | 541-46-8 | |
| PubChem CID | 10930 | |
| ChEMBL ID | CHEMBL171066 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 101.15 | ALogp: | 0.4 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 43.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 7 | QED Weighted: | 0.552 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.511 | MDCK Permeability: | 0.00009320 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.797 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.001 |
| 30% Bioavailability (F30%): | 0.001 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.999 | Plasma Protein Binding (PPB): | 28.31% |
| Volume Distribution (VD): | 0.978 | Fu: | 77.80% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.247 | CYP1A2-substrate: | 0.119 |
| CYP2C19-inhibitor: | 0.027 | CYP2C19-substrate: | 0.448 |
| CYP2C9-inhibitor: | 0.021 | CYP2C9-substrate: | 0.534 |
| CYP2D6-inhibitor: | 0.008 | CYP2D6-substrate: | 0.203 |
| CYP3A4-inhibitor: | 0.008 | CYP3A4-substrate: | 0.175 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.237 | Half-life (T1/2): | 0.386 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.016 | Human Hepatotoxicity (H-HT): | 0.237 |
| Drug-inuced Liver Injury (DILI): | 0.058 | AMES Toxicity: | 0.042 |
| Rat Oral Acute Toxicity: | 0.037 | Maximum Recommended Daily Dose: | 0.021 |
| Skin Sensitization: | 0.218 | Carcinogencity: | 0.054 |
| Eye Corrosion: | 0.028 | Eye Irritation: | 0.949 |
| Respiratory Toxicity: | 0.021 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000237 |  |
0.545 | D00WUF | 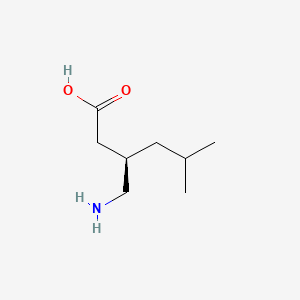 |
0.364 | ||
| ENC000351 | 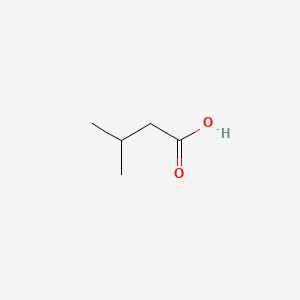 |
0.545 | D09PUL | 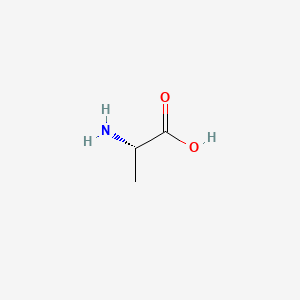 |
0.304 | ||
| ENC000682 |  |
0.500 | D07ZTO |  |
0.294 | ||
| ENC000685 | 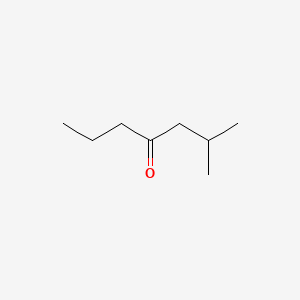 |
0.429 | D0ZK8H | 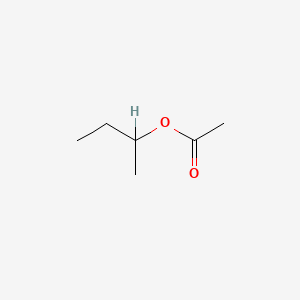 |
0.276 | ||
| ENC000241 | 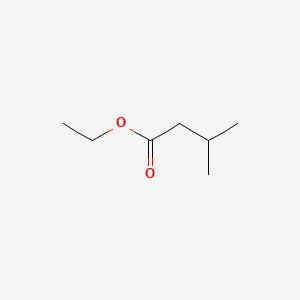 |
0.429 | D02XBW |  |
0.250 | ||
| ENC000445 | 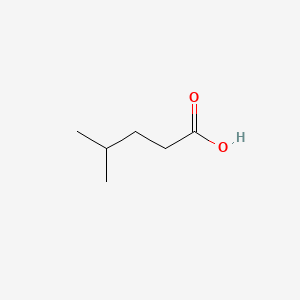 |
0.370 | D08QGD | 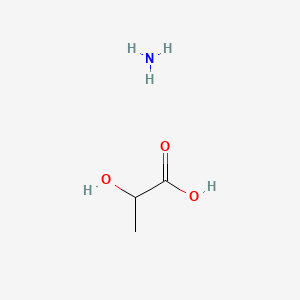 |
0.240 | ||
| ENC000246 |  |
0.370 | D00ZOF |  |
0.240 | ||
| ENC000149 | 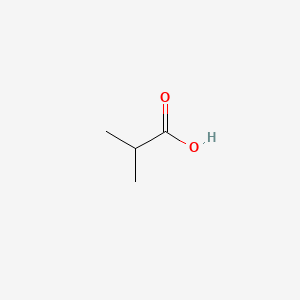 |
0.364 | D0U7BW | 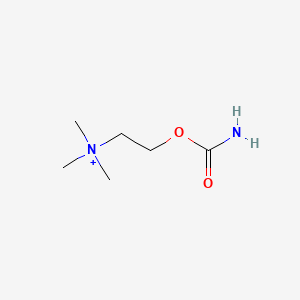 |
0.235 | ||
| ENC000397 |  |
0.364 | D01JIA | 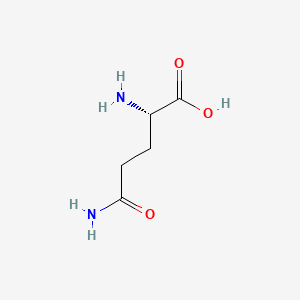 |
0.235 | ||
| ENC001734 |  |
0.364 | D08HZC | 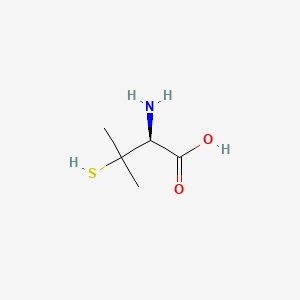 |
0.226 | ||