NPs Basic Information

|
Name |
2',5'-Dihydroxyacetophenone
|
| Molecular Formula | C8H8O3 | |
| IUPAC Name* |
1-(2,5-dihydroxyphenyl)ethanone
|
|
| SMILES |
CC(=O)C1=C(C=CC(=C1)O)O
|
|
| InChI |
InChI=1S/C8H8O3/c1-5(9)7-4-6(10)2-3-8(7)11/h2-4,10-11H,1H3
|
|
| InChIKey |
WLDWSGZHNBANIO-UHFFFAOYSA-N
|
|
| Synonyms |
490-78-8; 2',5'-DIHYDROXYACETOPHENONE; 1-(2,5-Dihydroxyphenyl)ethanone; 2,5-Dihydroxyacetophenone; 2-Acetylhydroquinone; Quinacetophenone; Acetylhydroquinone; Ethanone, 1-(2,5-dihydroxyphenyl)-; 1-(2,5-dihydroxyphenyl)ethan-1-one; Acetylquinol; Acetophenone, 2',5'-dihydroxy-; MFCD00002343; 1-Acetyl-2,5-dihydroxybenzene; NSC 3759; 2,5-Dihydroxy-1-acetylbenzene; K1H7QH11ZH; ghl.PD_Mitscher_leg0.355; 2', 5'-Dihydroxyacetophenone; 2-Acetyl-1,4-dihydroxybenzene; 2,5-Dihydroxyphenyl methyl ketone; NSC-3759; 2',5'-Dihydroxy acetophenone; EINECS 207-716-4; UNII-K1H7QH11ZH; BRN 0637903; AI3-18221; NSC3759; 2-acetyl hydroquinone; 2-acetyl-hydroquinone; monoacetyl hydroquinone; 2-5-dihydroxyacetophenone; 2,5 -dihydroxyacetophenone; WLN: QR CQ BV1; 2,5-Dihydroxy acetophenone; Acetophenone,5'-dihydroxy-; 2`,5`-Dihydroxyacetophenone; Ethanone,5-dihydroxyphenyl)-; 4-08-00-01803 (Beilstein Handbook Reference); BIDD:ER0488; SCHEMBL108053; 2',5'-Dihydroxy-Acetophenone; CHEMBL2348532; DTXSID9060077; CHEBI:173647; ZINC164902; 1-(2,5-dihydroxyphenyl)-ethanone; 2-ACETYL-1,4-BENZENEDIOL; 2-ACETYLHYDROQUINONE [INCI]; ACT00481; ALBB-019961; AMY27936; STR02363; 1-(2,5-dihydroxy-phenyl)-ethanone; 2',5'-Dihydroxyacetophenone, 97%; BBL027378; s9374; STL199230; AKOS000120785; CCG-266221; CS-W001174; FS-3211; HY-W001174; 1-(2,5-Dihydroxyphenyl)ethanone, 9CI; s12309; AC-19701; SY011591; D1955; DHAP, 2-Acetylhydroquinone, Quinacetophenone; FT-0610380; EN300-17127; A827653; AE-641/00693054; Q-200196; Q27281823; Z56889030; F3139-0239; 2 inverted exclamation mark ,5 inverted exclamation mark -Dihydroxyacetophenone
|
|
| CAS | 490-78-8 | |
| PubChem CID | 10279 | |
| ChEMBL ID | CHEMBL2348532 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 152.15 | ALogp: | 1.7 |
| HBD: | 2 | HBA: | 3 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 57.5 | Aromatic Rings: | 1 |
| Heavy Atoms: | 11 | QED Weighted: | 0.476 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.572 | MDCK Permeability: | 0.00001420 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.021 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.176 |
| 30% Bioavailability (F30%): | 0.174 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.088 | Plasma Protein Binding (PPB): | 71.30% |
| Volume Distribution (VD): | 0.792 | Fu: | 29.87% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.858 | CYP1A2-substrate: | 0.534 |
| CYP2C19-inhibitor: | 0.236 | CYP2C19-substrate: | 0.062 |
| CYP2C9-inhibitor: | 0.178 | CYP2C9-substrate: | 0.895 |
| CYP2D6-inhibitor: | 0.321 | CYP2D6-substrate: | 0.725 |
| CYP3A4-inhibitor: | 0.241 | CYP3A4-substrate: | 0.205 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 13.836 | Half-life (T1/2): | 0.897 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.024 | Human Hepatotoxicity (H-HT): | 0.105 |
| Drug-inuced Liver Injury (DILI): | 0.232 | AMES Toxicity: | 0.206 |
| Rat Oral Acute Toxicity: | 0.193 | Maximum Recommended Daily Dose: | 0.057 |
| Skin Sensitization: | 0.521 | Carcinogencity: | 0.349 |
| Eye Corrosion: | 0.893 | Eye Irritation: | 0.982 |
| Respiratory Toxicity: | 0.423 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000097 | 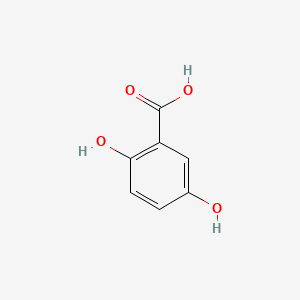 |
0.706 | D0C4YC | 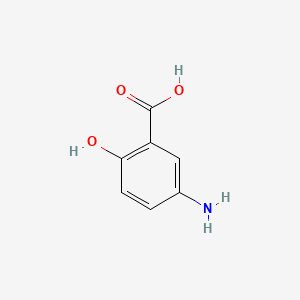 |
0.487 | ||
| ENC002913 |  |
0.641 | D01WJL | 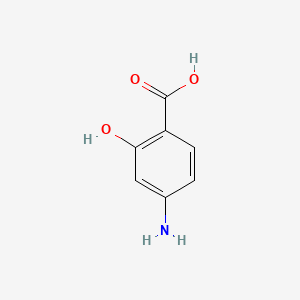 |
0.415 | ||
| ENC002464 |  |
0.610 | D07HBX |  |
0.400 | ||
| ENC003828 |  |
0.568 | D0V9EN | 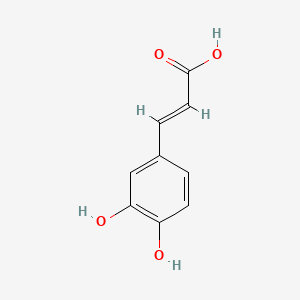 |
0.391 | ||
| ENC000069 | 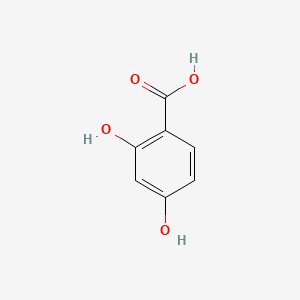 |
0.568 | D0BA6T | 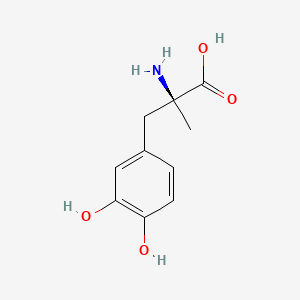 |
0.388 | ||
| ENC001108 |  |
0.526 | D0U0OT | 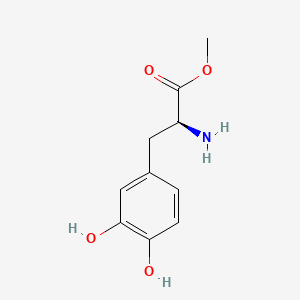 |
0.380 | ||
| ENC000696 |  |
0.514 | D0YF3X |  |
0.377 | ||
| ENC000986 |  |
0.488 | D08HVR | 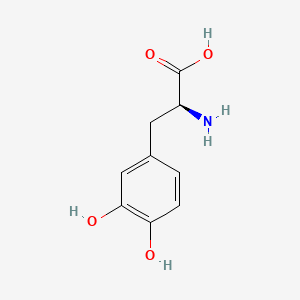 |
0.375 | ||
| ENC000103 | 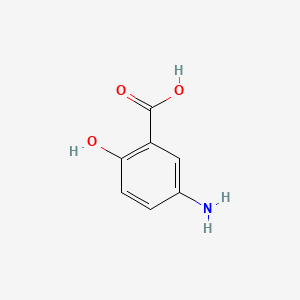 |
0.487 | D0U5QK | 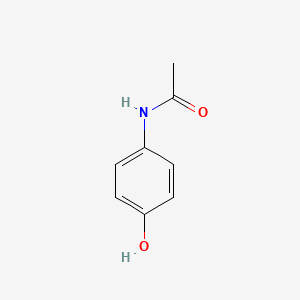 |
0.372 | ||
| ENC000690 | 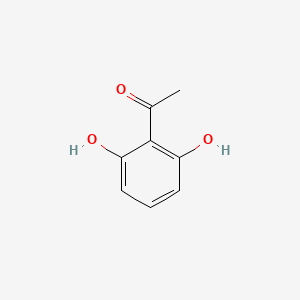 |
0.487 | D0S2BT | 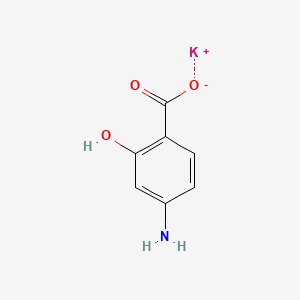 |
0.372 | ||