NPs Basic Information
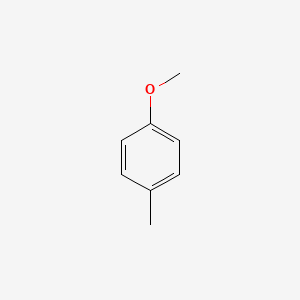
|
Name |
1-Methoxy-4-methylbenzene
|
| Molecular Formula | C8H10O | |
| IUPAC Name* |
1-methoxy-4-methylbenzene
|
|
| SMILES |
CC1=CC=C(C=C1)OC
|
|
| InChI |
InChI=1S/C8H10O/c1-7-3-5-8(9-2)6-4-7/h3-6H,1-2H3
|
|
| InChIKey |
CHLICZRVGGXEOD-UHFFFAOYSA-N
|
|
| Synonyms |
4-Methylanisole; 1-METHOXY-4-METHYLBENZENE; 104-93-8; 4-Methoxytoluene; p-Methylanisole; p-Methoxytoluene; Benzene, 1-methoxy-4-methyl-; p-methyl anisole; p-Cresol methyl ether; p-Cresyl methyl ether; Anisole, p-methyl-; 4-Methyl anisole; Methyl p-cresol; Methyl p-tolyl ether; p-Tolyl methyl ether; Methyl p-cresyl ether; 1-Methyl-4-methoxybenzene; 4-Methyl-1-methoxybenzene; 4-Methylphenol methyl ether; Methyl 4-methylphenyl ether; para-Methylanisole; Methyl-para-cresol; para-Methoxytoluene; Methyl p-methylphenyl ether; p-methylanisol; 4-methylanisol; para-Cresyl methyl ether; para-methyl anisole; 1-Methoxy-4-methyl-benzene; FEMA No. 2681; 4-Methylanizole; NSC 6254; para-Methyl anisol; 4-methylmethoxybenzene; 10FAI0OR9W; CHEMBL154155; CHEBI:89728; NSC-6254; Toluene, 4-methoxy-; FEMA Number 2681; PARA-CRESOL DEUTEROMETHYL ETHER; HSDB 5363; EINECS 203-253-7; UNII-10FAI0OR9W; AI3-07621; p- methylanisole; p-Methyl-Anisole; p-Methoxy toluene; 4-methoxy-toluene; MFCD00008413; VANADIUMGALLIDE; P- METHOXYTOLUENE; 4-Methylanisole, 99%; DSSTox_CID_6710; 1-methyoxy-4-methylbenzene; EC 203-253-7; DSSTox_RID_78191; DSSTox_GSID_26710; SCHEMBL12464; 4-CRESOL METHYL ETHER; WLN: 1OR D1; P-METHYLANISOLE [FHFI]; SCHEMBL2489775; P-METHYL ANISOLE [FCC]; PARA-CRESOL METHYL ETHER; DTXSID9026710; SCHEMBL12015216; cresyl methyl ether, methylanisol; FEMA 2681; AMY5180; NSC6254; ZINC1693359; 4-Methylanisole, analytical standard; Tox21_201081; BDBM50008555; STL268886; AKOS000121016; 4-Methylanisole, >=99%, FCC, FG; CS-W013551; HY-W012835; NCGC00248917-01; NCGC00258634-01; 1-METHOXY-4-METHYLBENZENE [HSDB]; BS-23266; CAS-104-93-8; FT-0618970; M0149; EN300-16112; W-108799; Q15726084; Z53834227; F0001-0094
|
|
| CAS | 104-93-8 | |
| PubChem CID | 7731 | |
| ChEMBL ID | CHEMBL154155 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 122.16 | ALogp: | 2.7 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 9.2 | Aromatic Rings: | 1 |
| Heavy Atoms: | 9 | QED Weighted: | 0.556 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.287 | MDCK Permeability: | 0.00002320 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.008 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.116 |
| 30% Bioavailability (F30%): | 0.335 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.885 | Plasma Protein Binding (PPB): | 87.22% |
| Volume Distribution (VD): | 2.234 | Fu: | 9.73% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.964 | CYP1A2-substrate: | 0.958 |
| CYP2C19-inhibitor: | 0.914 | CYP2C19-substrate: | 0.878 |
| CYP2C9-inhibitor: | 0.349 | CYP2C9-substrate: | 0.869 |
| CYP2D6-inhibitor: | 0.374 | CYP2D6-substrate: | 0.92 |
| CYP3A4-inhibitor: | 0.142 | CYP3A4-substrate: | 0.525 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.149 | Half-life (T1/2): | 0.677 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.088 | Human Hepatotoxicity (H-HT): | 0.09 |
| Drug-inuced Liver Injury (DILI): | 0.265 | AMES Toxicity: | 0.104 |
| Rat Oral Acute Toxicity: | 0.032 | Maximum Recommended Daily Dose: | 0.036 |
| Skin Sensitization: | 0.634 | Carcinogencity: | 0.674 |
| Eye Corrosion: | 0.975 | Eye Irritation: | 0.994 |
| Respiratory Toxicity: | 0.105 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000318 | 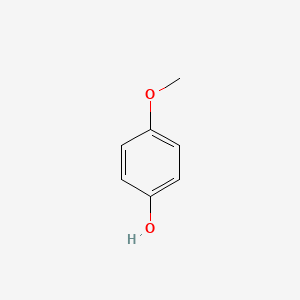 |
0.613 | D02DPU | 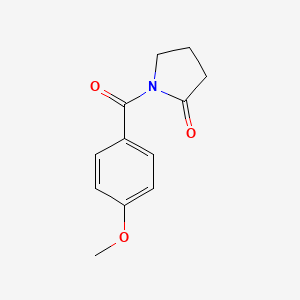 |
0.373 | ||
| ENC000201 |  |
0.571 | D0DJ1B |  |
0.333 | ||
| ENC000233 | 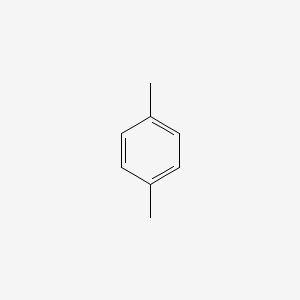 |
0.567 | D0P1UX | 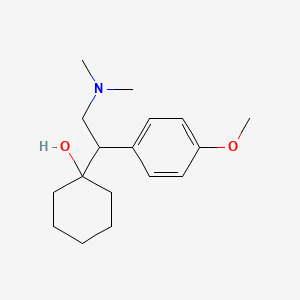 |
0.328 | ||
| ENC000223 |  |
0.559 | D05CKR | 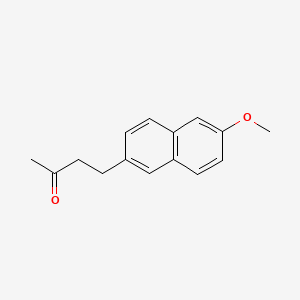 |
0.327 | ||
| ENC001460 |  |
0.556 | D09WKB | 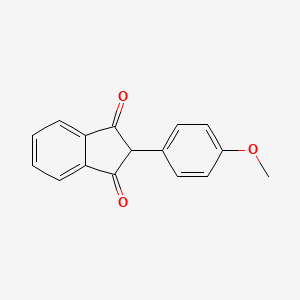 |
0.317 | ||
| ENC000298 |  |
0.526 | D0J5DC |  |
0.308 | ||
| ENC000086 | 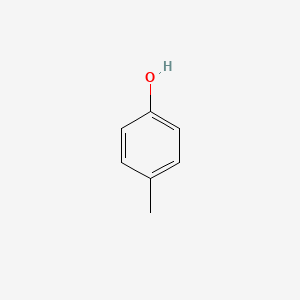 |
0.516 | D09GYT | 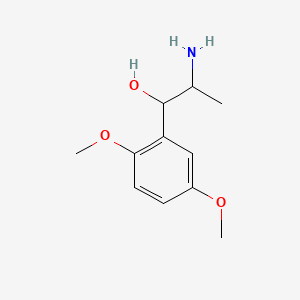 |
0.300 | ||
| ENC000310 |  |
0.514 | D08JZS |  |
0.299 | ||
| ENC000199 | 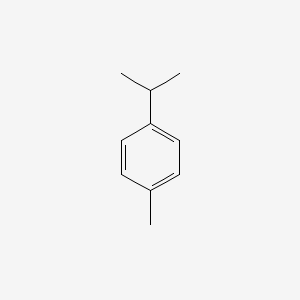 |
0.486 | D06OIV |  |
0.298 | ||
| ENC000638 |  |
0.475 | D0X0WU | 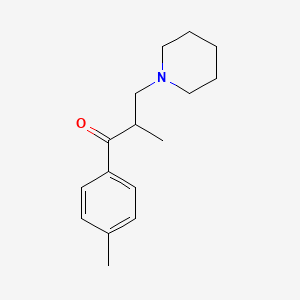 |
0.288 | ||