NPs Basic Information
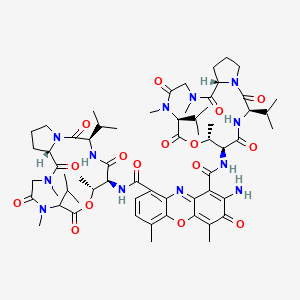
|
Name |
Actinomycin D
|
| Molecular Formula | C62H86N12O16 | |
| IUPAC Name* |
2-amino-4,6-dimethyl-3-oxo-1-N-[(3R,6S,7R,10S,16S)-7,11,14-trimethyl-2,5,9,12,15-pentaoxo-3,10-di(propan-2-yl)-8-oxa-1,4,11,14-tetrazabicyclo[14.3.0]nonadecan-6-yl]-9-N-[(3R,6S,7R,16S)-7,11,14-trimethyl-2,5,9,12,15-pentaoxo-3,10-di(propan-2-yl)-8-oxa-1,4,11,14-tetrazabicyclo[14.3.0]nonadecan-6-yl]phenoxazine-1,9-dicarboxamide
|
|
| SMILES |
C[C@@H]1[C@@H](C(=O)N[C@@H](C(=O)N2CCC[C@H]2C(=O)N(CC(=O)N([C@H](C(=O)O1)C(C)C)C)C)C(C)C)NC(=O)C3=C(C(=O)C(=C4C3=NC5=C(C=CC(=C5O4)C)C(=O)N[C@H]6[C@H](OC(=O)C(N(C(=O)CN(C(=O)[C@@H]7CCCN7C(=O)[C@H](NC6=O)C(C)C)C)C)C(C)C)C)C)N
|
|
| InChI |
InChI=1S/C62H86N12O16/c1-27(2)42-59(84)73-23-17-19-36(73)57(82)69(13)25-38(75)71(15)48(29(5)6)61(86)88-33(11)44(55(80)65-42)67-53(78)35-22-21-31(9)51-46(35)64-47-40(41(63)50(77)32(10)52(47)90-51)54(79)68-45-34(12)89-62(87)49(30(7)8)72(16)39(76)26-70(14)58(83)37-20-18-24-74(37)60(85)43(28(3)4)66-56(45)81/h21-22,27-30,33-34,36-37,42-45,48-49H,17-20,23-26,63H2,1-16H3,(H,65,80)(H,66,81)(H,67,78)(H,68,79)/t33-,34-,36+,37+,42-,43-,44+,45+,48?,49+/m1/s1
|
|
| InChIKey |
RJURFGZVJUQBHK-HUZQGMAJSA-N
|
|
| Synonyms |
actinomycin D; 50-76-0
|
|
| CAS | 50-76-0 | |
| PubChem CID | 131954673 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 1255.4 | ALogp: | 3.8 |
| HBD: | 5 | HBA: | 18 |
| Rotatable Bonds: | 8 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 356.0 | Aromatic Rings: | 7 |
| Heavy Atoms: | 90 | QED Weighted: | 0.12 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.607 | MDCK Permeability: | 0.00001560 |
| Pgp-inhibitor: | 0.528 | Pgp-substrate: | 1 |
| Human Intestinal Absorption (HIA): | 0.889 | 20% Bioavailability (F20%): | 0.092 |
| 30% Bioavailability (F30%): | 0.66 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.094 | Plasma Protein Binding (PPB): | 91.10% |
| Volume Distribution (VD): | 0.488 | Fu: | 10.23% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0 | CYP1A2-substrate: | 0.01 |
| CYP2C19-inhibitor: | 0.045 | CYP2C19-substrate: | 0.059 |
| CYP2C9-inhibitor: | 0.125 | CYP2C9-substrate: | 0.058 |
| CYP2D6-inhibitor: | 0.004 | CYP2D6-substrate: | 0.078 |
| CYP3A4-inhibitor: | 0.557 | CYP3A4-substrate: | 0.908 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 3.731 | Half-life (T1/2): | 0.075 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.001 | Human Hepatotoxicity (H-HT): | 0.986 |
| Drug-inuced Liver Injury (DILI): | 0.993 | AMES Toxicity: | 0.003 |
| Rat Oral Acute Toxicity: | 0.013 | Maximum Recommended Daily Dose: | 0.614 |
| Skin Sensitization: | 0.379 | Carcinogencity: | 0.009 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.001 |
| Respiratory Toxicity: | 0.034 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000998 |  |
0.920 | D0P8IV |  |
1.000 | ||
| ENC005526 |  |
0.781 | D07XGH | 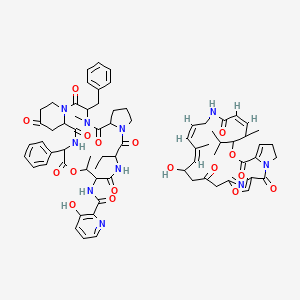 |
0.329 | ||
| ENC000948 | 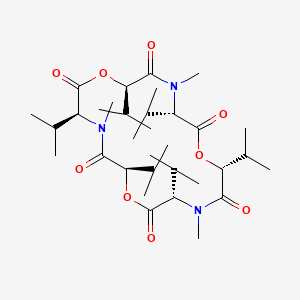 |
0.297 | D05MNW | 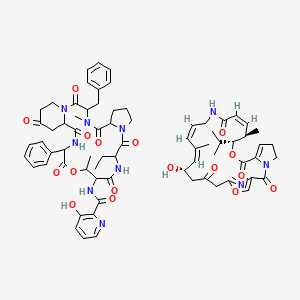 |
0.329 | ||
| ENC002129 | 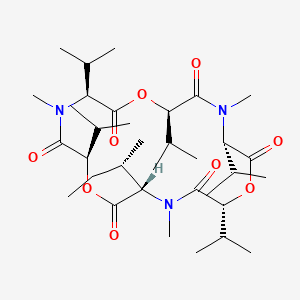 |
0.293 | D0O3YF |  |
0.319 | ||
| ENC002857 | 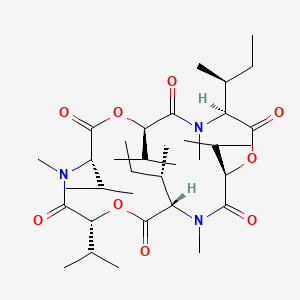 |
0.281 | D0L9HX | 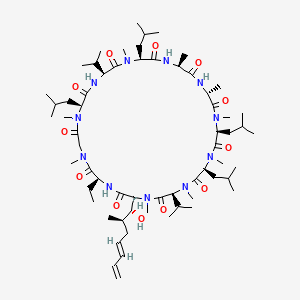 |
0.316 | ||
| ENC003559 | 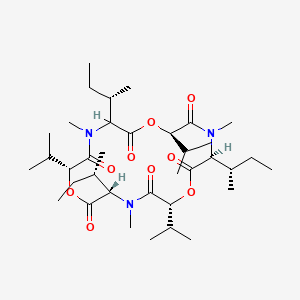 |
0.265 | D0J7XL | 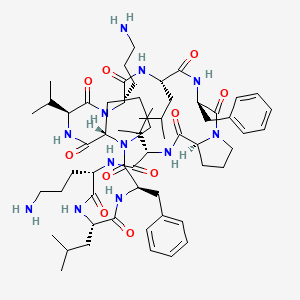 |
0.305 | ||
| ENC005563 | 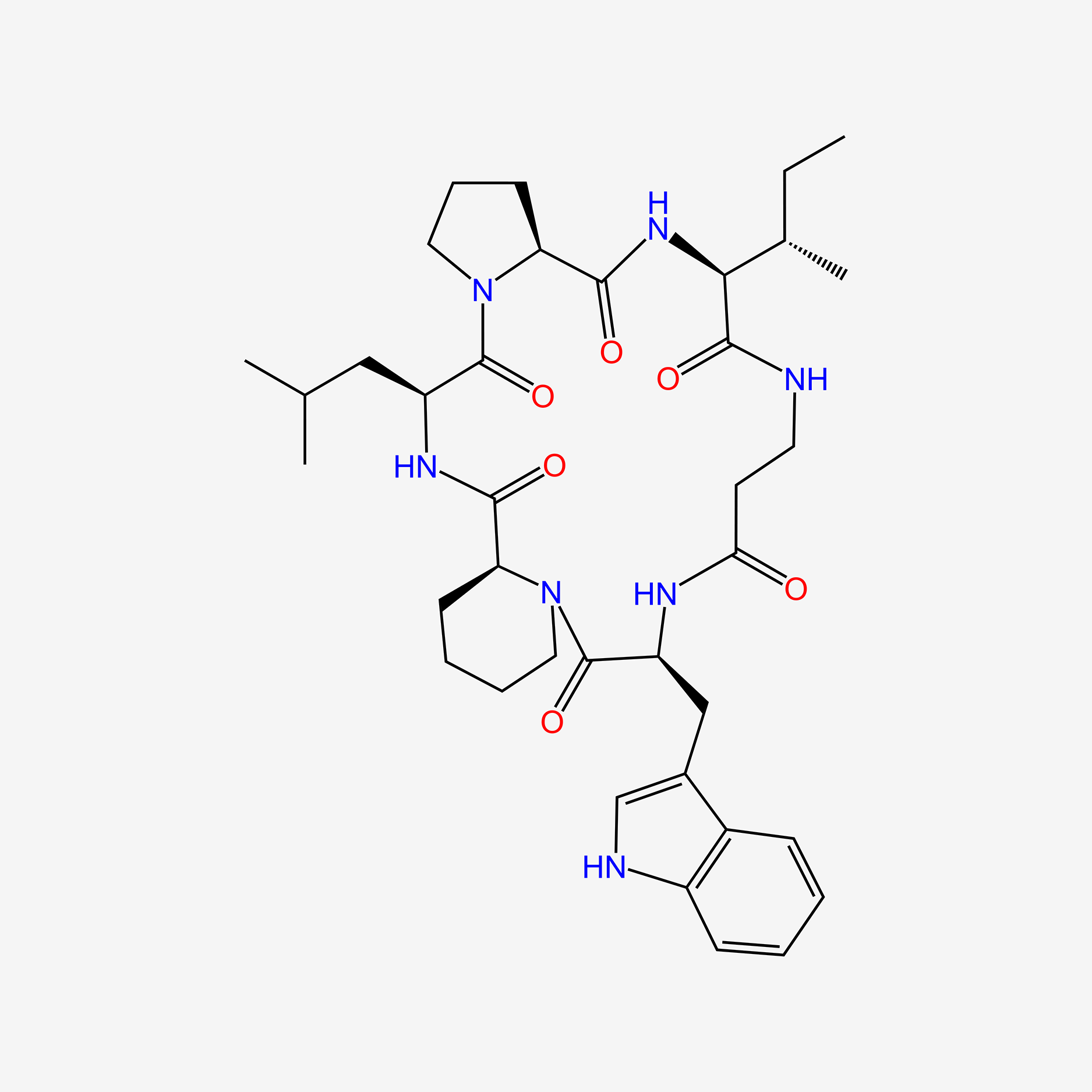 |
0.250 | D0E2OU | 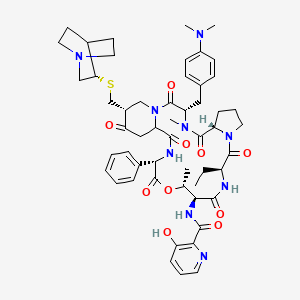 |
0.290 | ||
| ENC002483 |  |
0.245 | D06TOE | 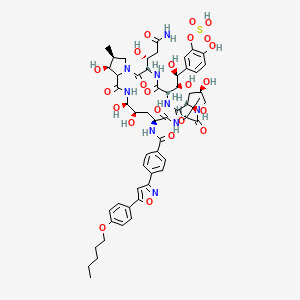 |
0.220 | ||
| ENC003706 | 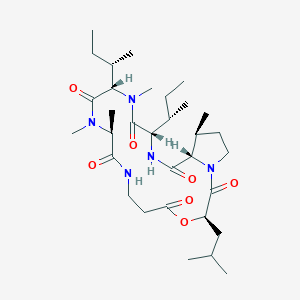 |
0.245 | D00GNJ |  |
0.219 | ||
| ENC003645 |  |
0.244 | D06WKA |  |
0.218 | ||