NPs Basic Information
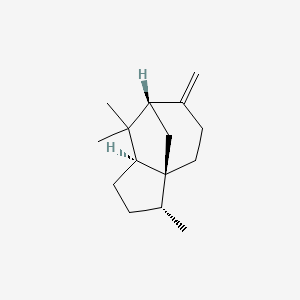
|
Name |
beta-Cedrene
|
| Molecular Formula | C15H24 | |
| IUPAC Name* |
(1S,2R,5S,7S)-2,6,6-trimethyl-8-methylidenetricyclo[5.3.1.01,5]undecane
|
|
| SMILES |
C[C@@H]1CC[C@@H]2[C@]13CCC(=C)[C@H](C3)C2(C)C
|
|
| InChI |
InChI=1S/C15H24/c1-10-7-8-15-9-12(10)14(3,4)13(15)6-5-11(15)2/h11-13H,1,5-9H2,2-4H3/t11-,12+,13+,15+/m1/s1
|
|
| InChIKey |
DYLPEFGBWGEFBB-OSFYFWSMSA-N
|
|
| Synonyms |
beta-Cedrene; 546-28-1; (+)-beta-Cedrene; .beta.-Cedrene; (+)-.beta.-Cedrene; 6QL7ERD5Q1; 1H-3a,7-Methanoazulene, octahydro-3,8,8-trimethyl-6-methylene-, (3R,3aS,7S,8aS)-; (+)-beta-Funebrene; Cedr-8(15)-ene; (1S,2R,5S,7S)-2,6,6-trimethyl-8-methylidenetricyclo[5.3.1.01,5]undecane; (3R,3aS,7S,8aS)-3,8,8-Trimethyl-6-methyleneoctahydro-1H-3a,7-methanoazulene; 1H-3a,7-Methanoazulene,octahydro-3,8,8-trimethyl-6-methylene-,(3R,3aS,7S,8aS)-; 1H-3a,7-Methanoazulene, octahydro-3,8,8-trimethyl-6-methylene-, [3R-(3.alpha.,3a.beta.,7.beta.,8a.alpha.)]-; 1H-3A,7-METHANOAZULENE, OCTAHYDRO-3,8,8-TRIMETHYL-6-METHYLENE-, (3R-(3.ALPHA.,3A.BETA.,7.BETA.,8A.ALPHA.))-; EINECS 208-898-8; (+)- beta -Cedrene; UNII-6QL7ERD5Q1; DTXSID40883435; MFCD00270433; ZINC59778864; AKOS015896901; 1H-3a,7-Methanoazulene, octahydro-3,8,8-trimethyl-6-methylene-, (3R-(3alpha,3abeta,7beta,8aalpha))-; (+)-beta-Cedrene, >=95.0% (sum of enantiomers, GC); (3R-(3alpha,3Abeta,7beta,8aalpha))-octahydro-3,8,8-trimethyl-6-methylene-1H-3a,7-methanoazulene
|
|
| CAS | 546-28-1 | |
| PubChem CID | 11106485 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 204.35 | ALogp: | 4.9 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 3 |
| Heavy Atoms: | 15 | QED Weighted: | 0.491 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.573 | MDCK Permeability: | 0.00001850 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.923 |
| 30% Bioavailability (F30%): | 0.864 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.228 | Plasma Protein Binding (PPB): | 82.71% |
| Volume Distribution (VD): | 0.809 | Fu: | 16.80% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.372 | CYP1A2-substrate: | 0.299 |
| CYP2C19-inhibitor: | 0.254 | CYP2C19-substrate: | 0.851 |
| CYP2C9-inhibitor: | 0.334 | CYP2C9-substrate: | 0.265 |
| CYP2D6-inhibitor: | 0.054 | CYP2D6-substrate: | 0.691 |
| CYP3A4-inhibitor: | 0.263 | CYP3A4-substrate: | 0.26 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 13.3 | Half-life (T1/2): | 0.061 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.021 | Human Hepatotoxicity (H-HT): | 0.244 |
| Drug-inuced Liver Injury (DILI): | 0.234 | AMES Toxicity: | 0.007 |
| Rat Oral Acute Toxicity: | 0.715 | Maximum Recommended Daily Dose: | 0.956 |
| Skin Sensitization: | 0.124 | Carcinogencity: | 0.472 |
| Eye Corrosion: | 0.92 | Eye Irritation: | 0.925 |
| Respiratory Toxicity: | 0.953 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC003109 | 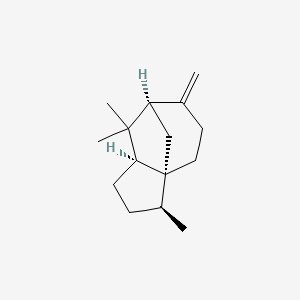 |
1.000 | D04VIS | 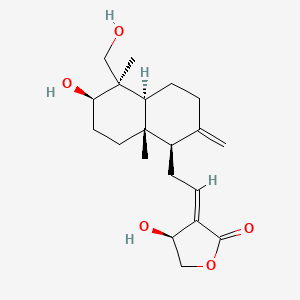 |
0.267 | ||
| ENC003097 | 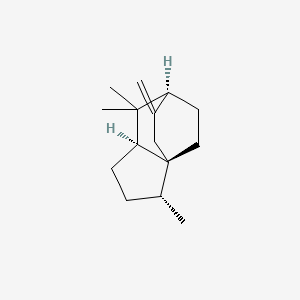 |
0.673 | D0D2VS | 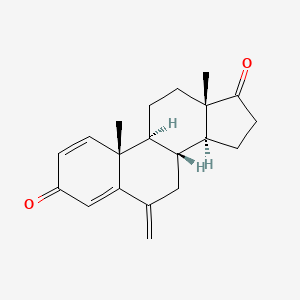 |
0.259 | ||
| ENC001831 |  |
0.577 | D04SFH | 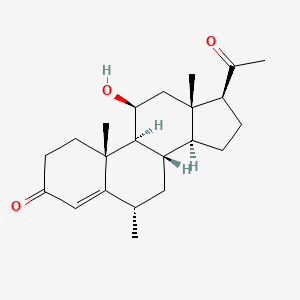 |
0.253 | ||
| ENC002998 | 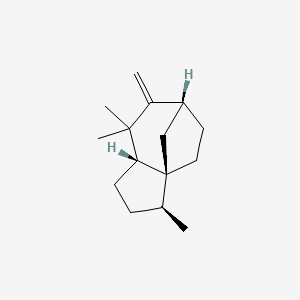 |
0.577 | D0H1QY |  |
0.250 | ||
| ENC001893 | 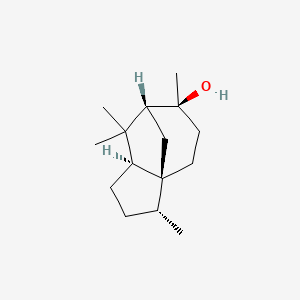 |
0.527 | D0L2LS | 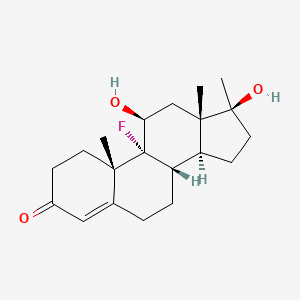 |
0.247 | ||
| ENC001172 | 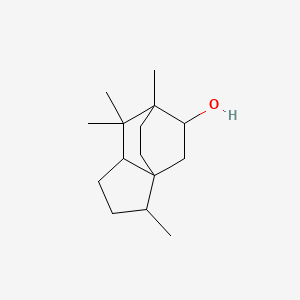 |
0.474 | D0Z1XD | 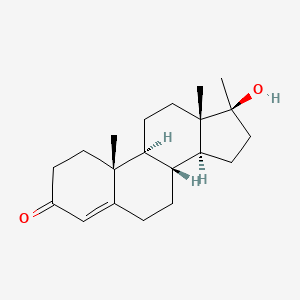 |
0.244 | ||
| ENC003477 | 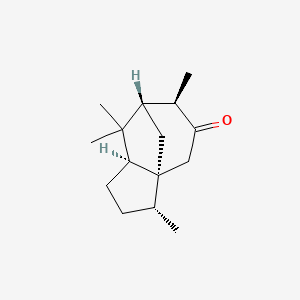 |
0.474 | D0I2SD |  |
0.239 | ||
| ENC003096 |  |
0.474 | D0V8HA |  |
0.237 | ||
| ENC003215 |  |
0.464 | D0F1UL |  |
0.235 | ||
| ENC002267 |  |
0.458 | D0G8BV |  |
0.235 | ||