NPs Basic Information
|
Name |
Linoelaidic acid
|
| Molecular Formula | C18H32O2 | |
| IUPAC Name* |
(9E,12E)-octadeca-9,12-dienoic acid
|
|
| SMILES |
CCCCC/C=C/C/C=C/CCCCCCCC(=O)O
|
|
| InChI |
InChI=1S/C18H32O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18(19)20/h6-7,9-10H,2-5,8,11-17H2,1H3,(H,19,20)/b7-6+,10-9+
|
|
| InChIKey |
OYHQOLUKZRVURQ-AVQMFFATSA-N
|
|
| Synonyms |
Linoelaidic acid; 506-21-8; (9E,12E)-octadeca-9,12-dienoic acid; LINOLELAIDIC ACID; 9E,12E-octadecadienoic acid; 9,12-Octadecadienoic acid; Linelaidic acid; 9,12-Octadecadienoic acid, (9E,12E)-; trans-9,trans-12-Linoleic acid; trans-9,trans-12-Octadecadienoic acid; (9E,12E)-9,12-Octadecadienoic acid; octadeca-9,12-dienoic acid; Linolsaure; 7552P0K6PN; LINOELAIDICACID; 9E,12E-octadecadienoate; SCHEMBL120856; 18:2, n-6,9 all-trans; C18:2, n-6,9 all-trans; SCHEMBL18133791; CHEBI:75108; CHEBI:92157; TRANS,TRANS-LINOLEIC ACID; DTXSID50897508; HMS3649F11; ZINC3802188; (9E,12E)-9,12-Octadecadienoate; BBL027461; LMFA01030123; STK801960; Linolelaidic acid, analytical standard; AKOS015892939; trans,trans-9,12-Octadecadienoic Acid; (9E,12E)-octadeca-9,12-dienoicacid; NCGC00344326-02; TRANS-9-TRANS-12-LINOLEIC ACID; LS-14679; VS-08542; Linolelaidic acid 10 microg/mL in Methanol; D75304; EN300-378340; TRANS-.DELTA.9,12-OCTADECADIENOIC ACID; EN300-6497943; L001067; SR-01000946651; Q2823277; SR-01000946651-1; W-203254
|
|
| CAS | 506-21-8 | |
| PubChem CID | 5282457 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 280.4 | ALogp: | 6.8 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 14 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 37.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 20 | QED Weighted: | 0.314 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.108 | MDCK Permeability: | 0.00003720 |
| Pgp-inhibitor: | 0.002 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.041 | 20% Bioavailability (F20%): | 0.97 |
| 30% Bioavailability (F30%): | 0.846 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.012 | Plasma Protein Binding (PPB): | 100.21% |
| Volume Distribution (VD): | 0.463 | Fu: | 0.53% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.243 | CYP1A2-substrate: | 0.185 |
| CYP2C19-inhibitor: | 0.12 | CYP2C19-substrate: | 0.063 |
| CYP2C9-inhibitor: | 0.369 | CYP2C9-substrate: | 0.994 |
| CYP2D6-inhibitor: | 0.078 | CYP2D6-substrate: | 0.383 |
| CYP3A4-inhibitor: | 0.081 | CYP3A4-substrate: | 0.021 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.208 | Half-life (T1/2): | 0.858 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.038 | Human Hepatotoxicity (H-HT): | 0.189 |
| Drug-inuced Liver Injury (DILI): | 0.01 | AMES Toxicity: | 0.002 |
| Rat Oral Acute Toxicity: | 0.01 | Maximum Recommended Daily Dose: | 0.469 |
| Skin Sensitization: | 0.947 | Carcinogencity: | 0.053 |
| Eye Corrosion: | 0.962 | Eye Irritation: | 0.97 |
| Respiratory Toxicity: | 0.675 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
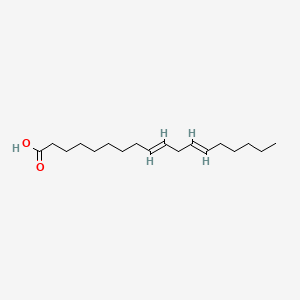
| ENC001535 |  |
1.000 | D0O1TC |  |
0.846 | ||
| ENC001920 |  |
0.839 | D0UE9X | 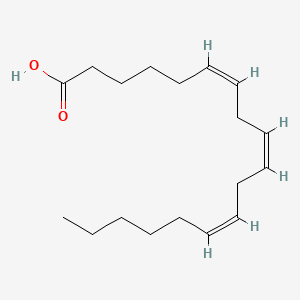 |
0.754 | ||
| ENC001099 | 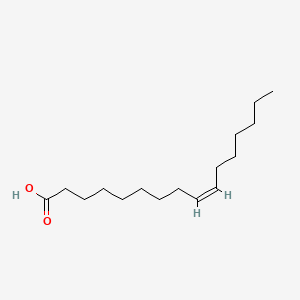 |
0.831 | D0O1PH |  |
0.662 | ||
| ENC001605 | 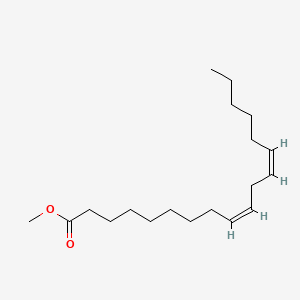 |
0.800 | D0OR6A | 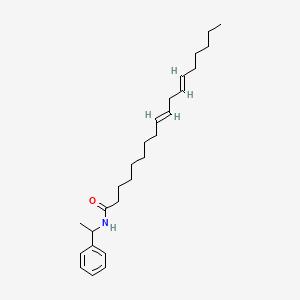 |
0.605 | ||
| ENC001583 | 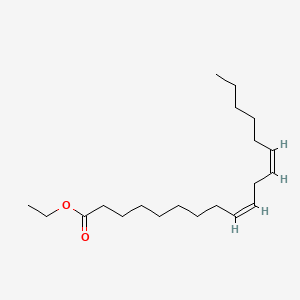 |
0.765 | D0Z5BC | 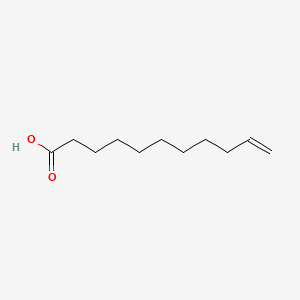 |
0.476 | ||
| ENC001554 | 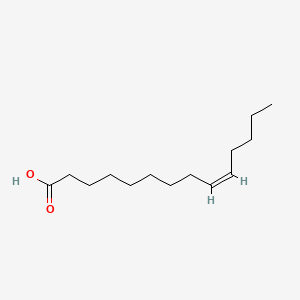 |
0.759 | D09SRR | 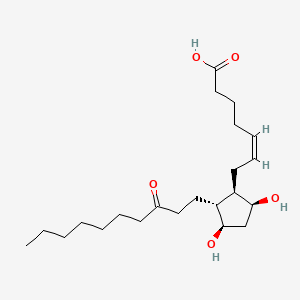 |
0.451 | ||
| ENC001544 | 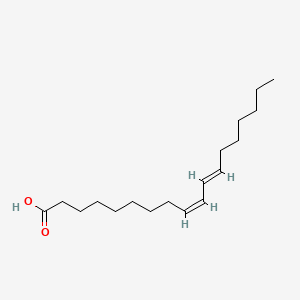 |
0.754 | D0XN8C | 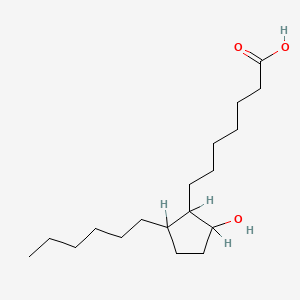 |
0.450 | ||
| ENC001555 | 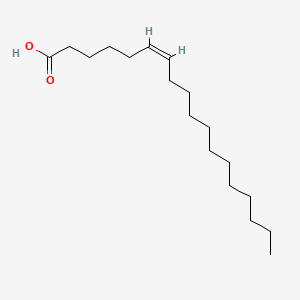 |
0.754 | D07ILQ |  |
0.393 | ||
| ENC001714 | 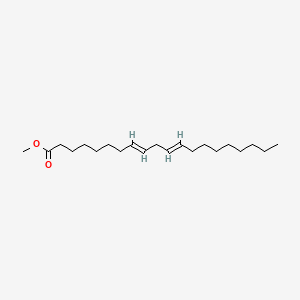 |
0.732 | D0I4DQ |  |
0.385 | ||
| ENC001715 |  |
0.707 | D0E4WR |  |
0.373 | ||