NPs Basic Information
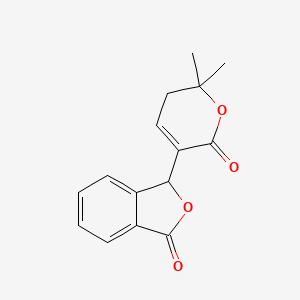
|
Name |
Catalpalactone
|
| Molecular Formula | C15H14O4 | |
| IUPAC Name* |
3-(2,2-dimethyl-6-oxo-3H-pyran-5-yl)-3H-2-benzofuran-1-one
|
|
| SMILES |
CC1(CC=C(C(=O)O1)C2C3=CC=CC=C3C(=O)O2)C
|
|
| InChI |
InChI=1S/C15H14O4/c1-15(2)8-7-11(14(17)19-15)12-9-5-3-4-6-10(9)13(16)18-12/h3-7,12H,8H2,1-2H3
|
|
| InChIKey |
GFYSRANGENPXDF-UHFFFAOYSA-N
|
|
| Synonyms |
Catalpalactone; 1585-68-8; 3-(2,2-dimethyl-6-oxo-3H-pyran-5-yl)-3H-2-benzofuran-1-one; P61GH0V29Z; CHEBI:80804; 1(3H)-Isobenzofuranone, 3-(5,6-dihydro-6,6-dimethyl-2-oxo-2H-pyran-3-yl)-; 3-(5,6-Dihydro-6,6-dimethyl-2-oxo-2H-pyran-3-yl)-1(3H)-isobenzofuranone; 3-(6,6-Dimethyl-2-oxo-5,6-dihydro-2H-pyran-3-yl)isobenzofuran-1(3H)-one; 1-Phthalanacetic acid, alpha-(3-hydroxy-3-methylbutylidene)-3-oxo-, delta-lactone; Phthalide, 3-(5,6-dihydro-6,6-dimethyl-2-oxo-2H-pyran-3-yl)-; UNII-P61GH0V29Z; starbld0005844; CHEMBL471809; DTXSID10928201; CATALPALACTONE, (+/-)-; AKOS032948471; C16929; Q27149847; 3-(6,6-Dimethyl-2-oxo-5,6-dihydro-2H-pyran-3-yl)-2-benzofuran-1(3H)-one; 1-PHTHALANACETIC ACID, .ALPHA.-(3-HYDROXY-3-METHYLBUTYLIDENE)-3-OXO-, .DELTA.-LACTONE; 133591-03-4
|
|
| CAS | 1585-68-8 | |
| PubChem CID | 3014018 | |
| ChEMBL ID | CHEMBL471809 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 258.27 | ALogp: | 2.4 |
| HBD: | 0 | HBA: | 4 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 52.6 | Aromatic Rings: | 3 |
| Heavy Atoms: | 19 | QED Weighted: | 0.725 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.65 | MDCK Permeability: | 0.00003510 |
| Pgp-inhibitor: | 0.064 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.454 |
| 30% Bioavailability (F30%): | 0.889 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.958 | Plasma Protein Binding (PPB): | 86.54% |
| Volume Distribution (VD): | 0.988 | Fu: | 17.53% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.41 | CYP1A2-substrate: | 0.525 |
| CYP2C19-inhibitor: | 0.764 | CYP2C19-substrate: | 0.605 |
| CYP2C9-inhibitor: | 0.482 | CYP2C9-substrate: | 0.507 |
| CYP2D6-inhibitor: | 0.022 | CYP2D6-substrate: | 0.482 |
| CYP3A4-inhibitor: | 0.422 | CYP3A4-substrate: | 0.283 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.747 | Half-life (T1/2): | 0.197 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.005 | Human Hepatotoxicity (H-HT): | 0.209 |
| Drug-inuced Liver Injury (DILI): | 0.849 | AMES Toxicity: | 0.029 |
| Rat Oral Acute Toxicity: | 0.45 | Maximum Recommended Daily Dose: | 0.015 |
| Skin Sensitization: | 0.216 | Carcinogencity: | 0.234 |
| Eye Corrosion: | 0.009 | Eye Irritation: | 0.207 |
| Respiratory Toxicity: | 0.202 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001031 | 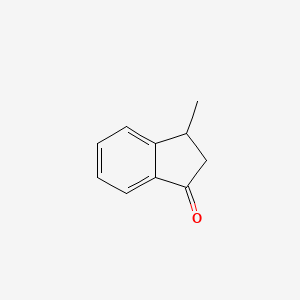 |
0.355 | D03GET | 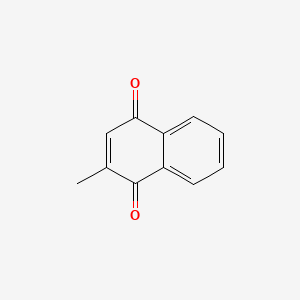 |
0.348 | ||
| ENC004792 | 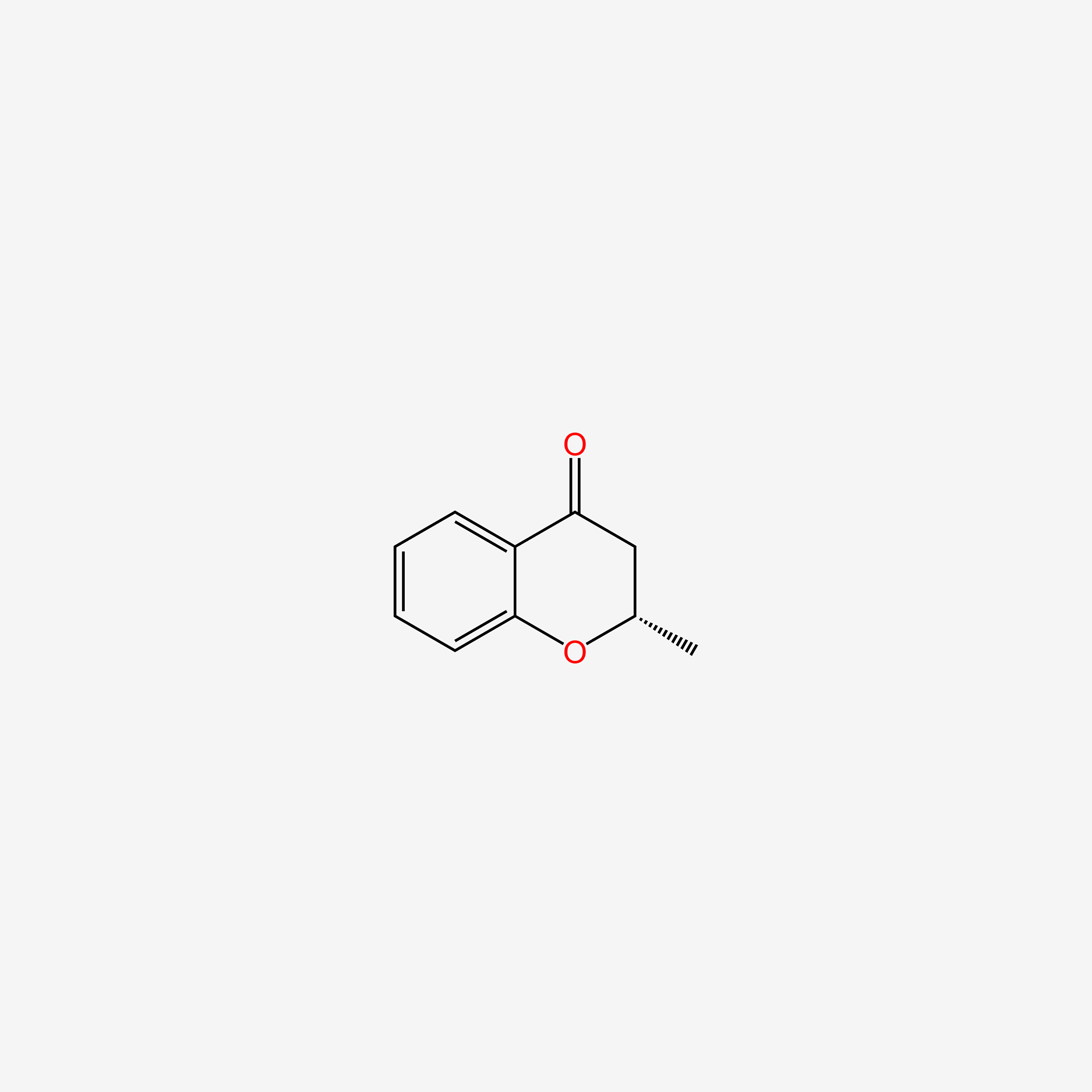 |
0.338 | D08CCE | 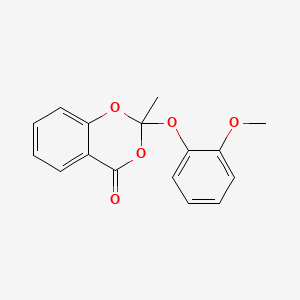 |
0.329 | ||
| ENC005855 | 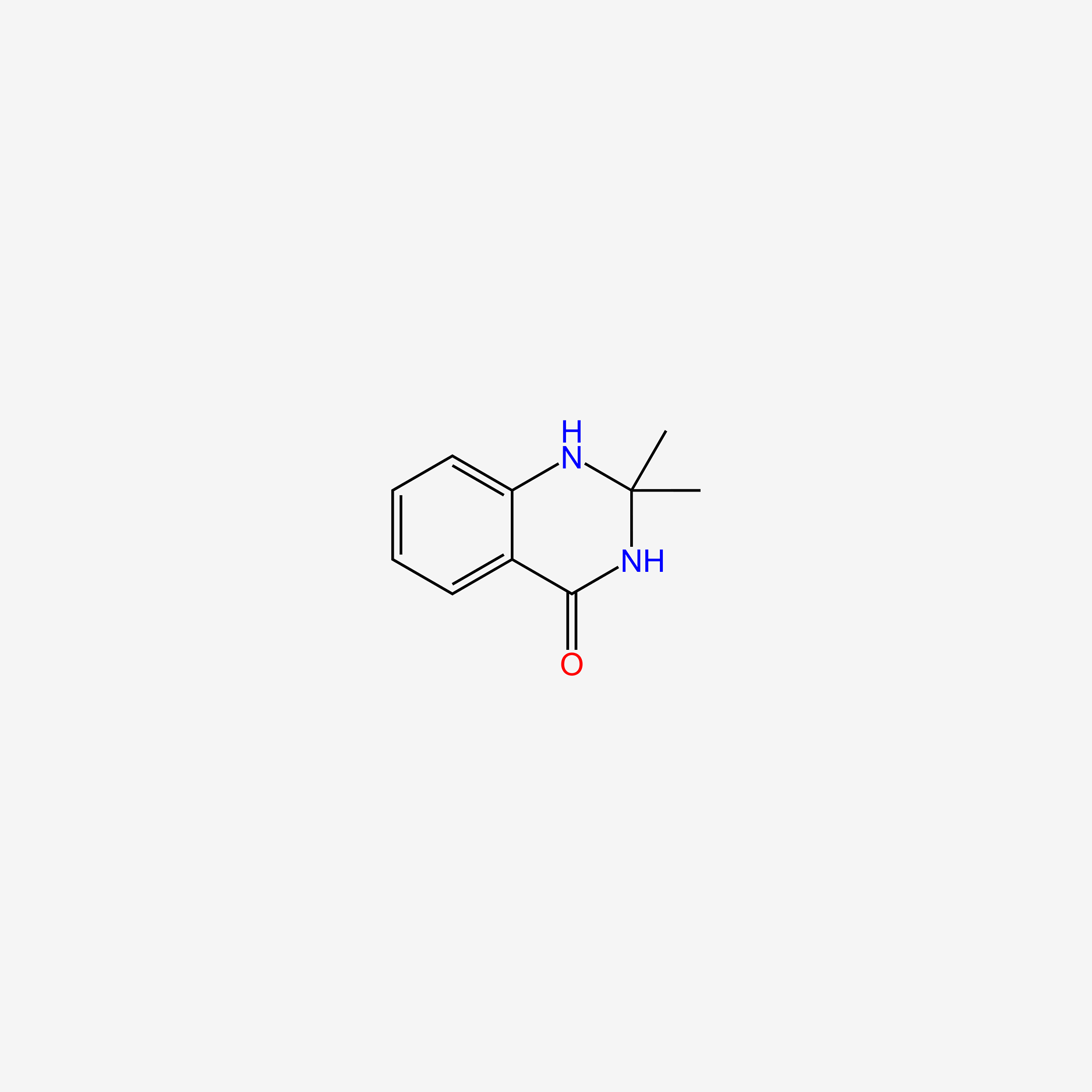 |
0.328 | D08EOD | 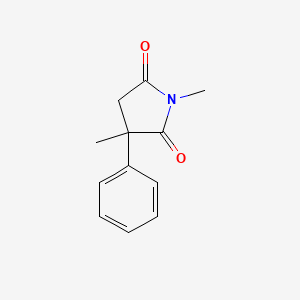 |
0.324 | ||
| ENC006142 | 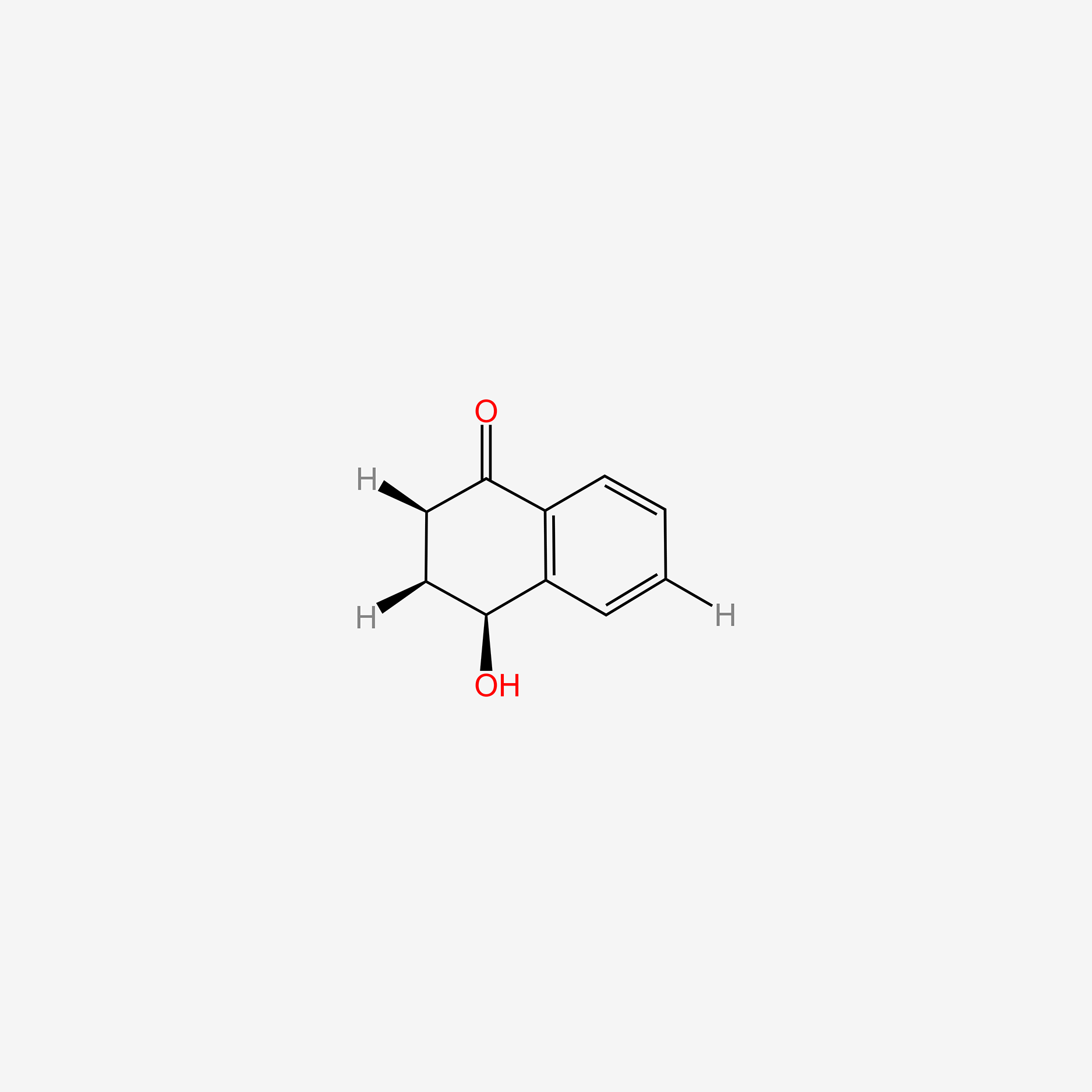 |
0.318 | D06BYV |  |
0.296 | ||
| ENC000973 |  |
0.316 | D08FTG |  |
0.291 | ||
| ENC000953 |  |
0.299 | D09WKB | 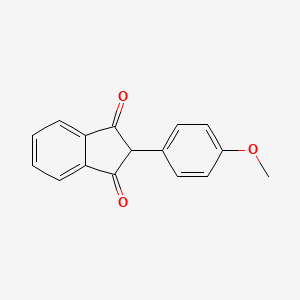 |
0.289 | ||
| ENC005244 | 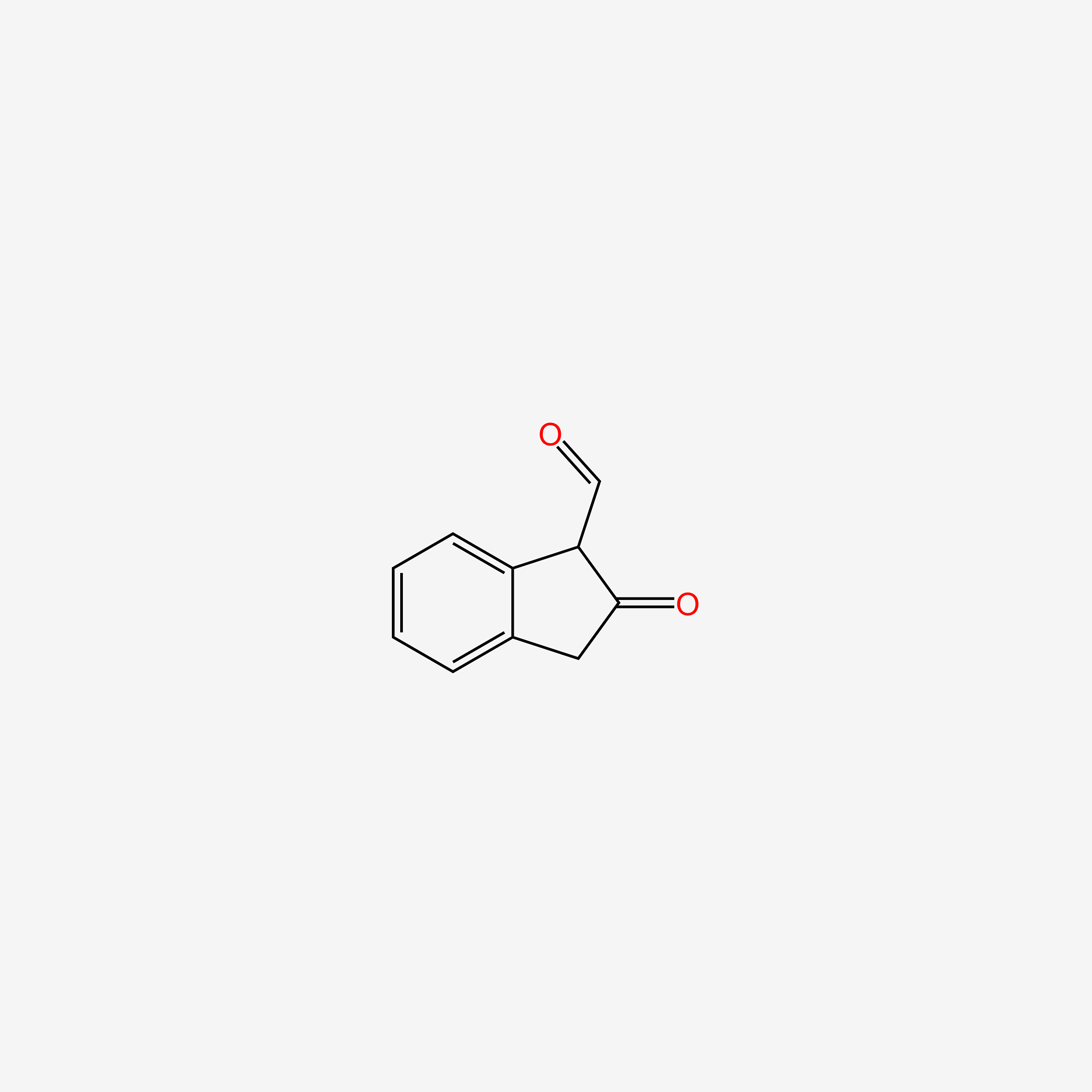 |
0.299 | D0D5GG | 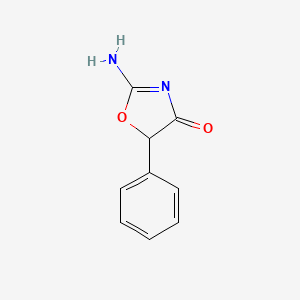 |
0.286 | ||
| ENC002130 | 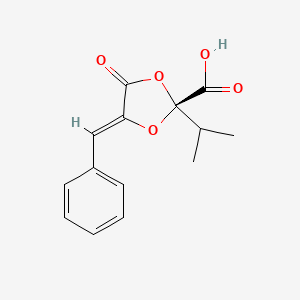 |
0.296 | D0U7GK | 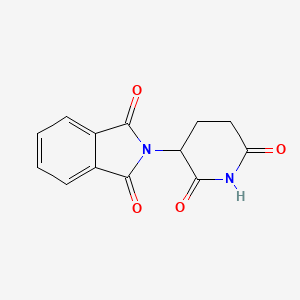 |
0.277 | ||
| ENC001372 | 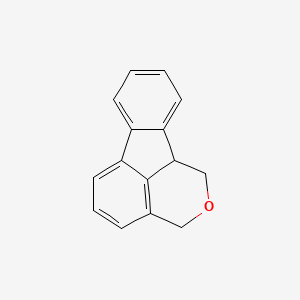 |
0.295 | D07RGW | 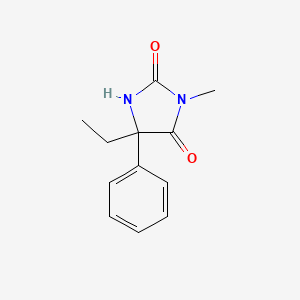 |
0.276 | ||
| ENC003914 | 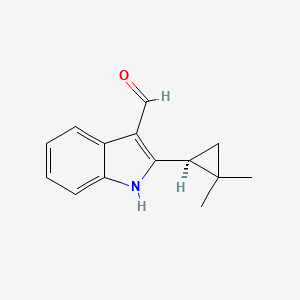 |
0.293 | D08UMH | 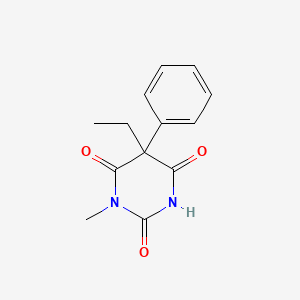 |
0.275 | ||