NPs Basic Information
|
Name |
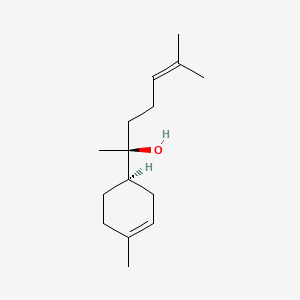
(+)-Epi-alpha-bisabolol
|
| Molecular Formula | C15H26O | |
| IUPAC Name* |
(2S)-6-methyl-2-[(1R)-4-methylcyclohex-3-en-1-yl]hept-5-en-2-ol
|
|
| SMILES |
CC1=CC[C@@H](CC1)[C@](C)(CCC=C(C)C)O
|
|
| InChI |
InChI=1S/C15H26O/c1-12(2)6-5-11-15(4,16)14-9-7-13(3)8-10-14/h6-7,14,16H,5,8-11H2,1-4H3/t14-,15-/m0/s1
|
|
| InChIKey |
RGZSQWQPBWRIAQ-GJZGRUSLSA-N
|
|
| Synonyms |
(+)-epi-alpha-bisabolol; alpha-Bisabolol; (+)-anymol; 76738-75-5; (2S)-6-methyl-2-[(1R)-4-methylcyclohex-3-en-1-yl]hept-5-en-2-ol; U799YDE8BR; 515-69-5; CHEBI:68658; (S)-6-Methyl-2-((R)-4-methylcyclohex-3-en-1-yl)hept-5-en-2-ol; 7-epi-.alpha.-Bisabolol; UNII-U799YDE8BR; alpha-Bisabolol, (+)-epi-; R-bisabolol; NSC606842; alpha -Bisabolol; (+)-epi-alpha-Bisabolol [MI]; epi-alpha-Bisabolol; CHEMBL519167; SCHEMBL1245143; ZINC968461; (+)-EPI-.ALPHA.-BISABOLOL; AKOS025295411; .ALPHA.-BISABOLOL, (+)-EPI-; 3-Cyclohexene-1-methanol, alpha,4-dimethyl-alpha-(4-methyl-3-penten-1-yl)-, (alphaS,1R)-; (+)-EPI-.ALPHA.-BISABOLOL [MI]; C20479; D88898; Q27137086; (2R)-6-methyl-2-[(1R)-4-methyl-1-cyclohex-3-enyl]hept-5-en-2-ol; 3-CYCLOHEXENE-1-METHANOL, .ALPHA.,4-DIMETHYL-.ALPHA.-(4-METHYL-3-PENTEN-1-YL)-, (.ALPHA.S,1R)-
|
|
| CAS | 76738-75-5 | |
| PubChem CID | 1201551 | |
| ChEMBL ID | CHEMBL519167 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 222.37 | ALogp: | 3.8 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 4 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 20.2 | Aromatic Rings: | 1 |
| Heavy Atoms: | 16 | QED Weighted: | 0.677 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.45 | MDCK Permeability: | 0.00001730 |
| Pgp-inhibitor: | 0.896 | Pgp-substrate: | 0.003 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.906 |
| 30% Bioavailability (F30%): | 0.104 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.091 | Plasma Protein Binding (PPB): | 93.58% |
| Volume Distribution (VD): | 2.439 | Fu: | 4.72% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.536 | CYP1A2-substrate: | 0.147 |
| CYP2C19-inhibitor: | 0.321 | CYP2C19-substrate: | 0.858 |
| CYP2C9-inhibitor: | 0.19 | CYP2C9-substrate: | 0.586 |
| CYP2D6-inhibitor: | 0.075 | CYP2D6-substrate: | 0.093 |
| CYP3A4-inhibitor: | 0.163 | CYP3A4-substrate: | 0.218 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 16.481 | Half-life (T1/2): | 0.198 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.032 | Human Hepatotoxicity (H-HT): | 0.825 |
| Drug-inuced Liver Injury (DILI): | 0.026 | AMES Toxicity: | 0.002 |
| Rat Oral Acute Toxicity: | 0.008 | Maximum Recommended Daily Dose: | 0.056 |
| Skin Sensitization: | 0.938 | Carcinogencity: | 0.4 |
| Eye Corrosion: | 0.784 | Eye Irritation: | 0.974 |
| Respiratory Toxicity: | 0.029 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005926 | 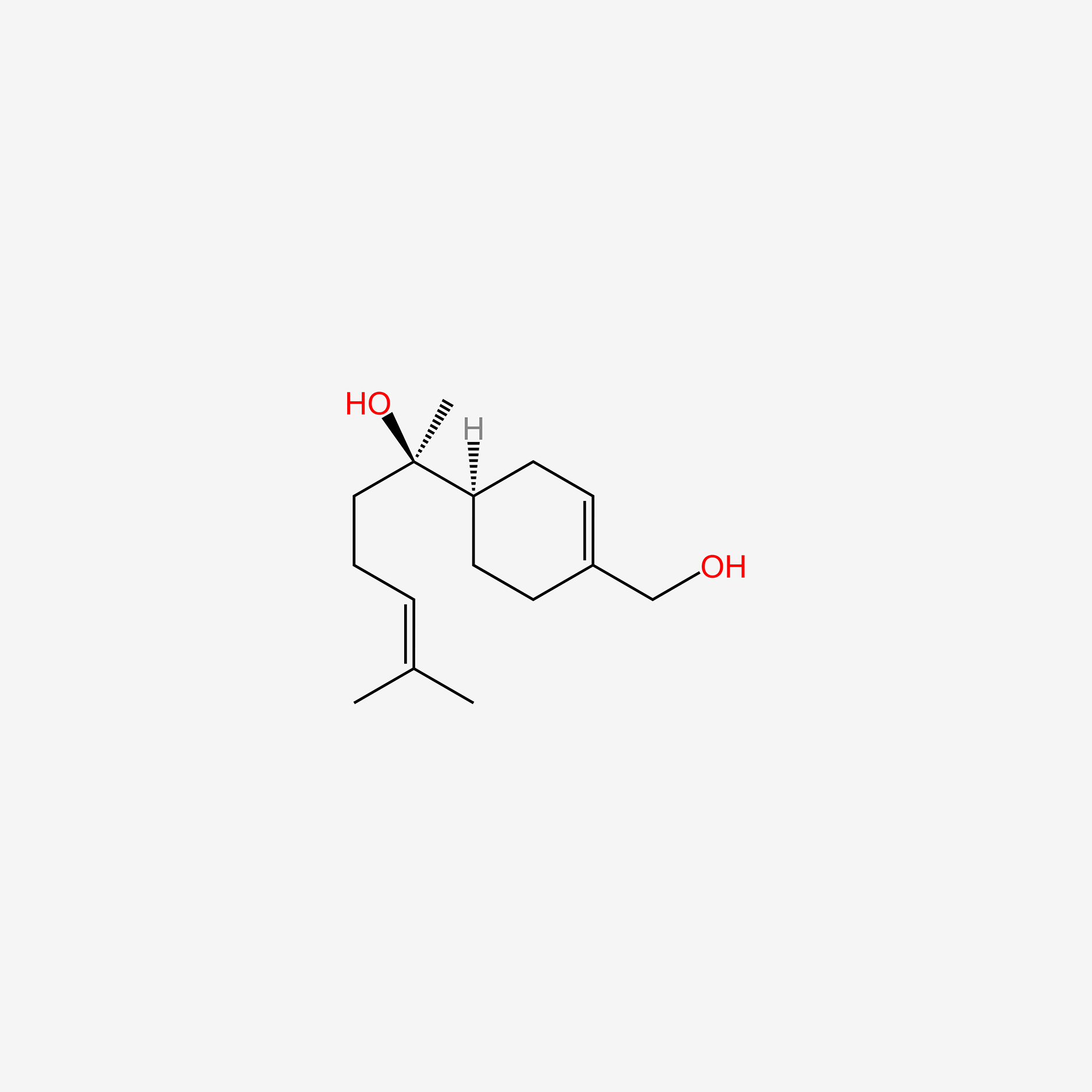 |
0.712 | D0M1PQ |  |
0.278 | ||
| ENC001812 | 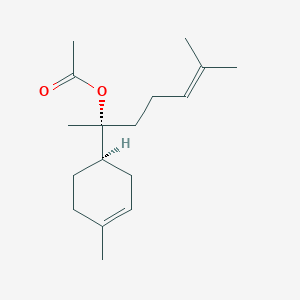 |
0.649 | D0W6DG | 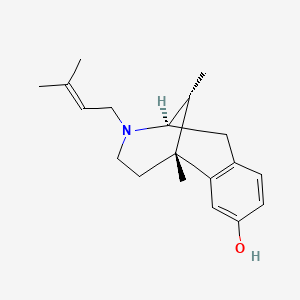 |
0.232 | ||
| ENC000511 | 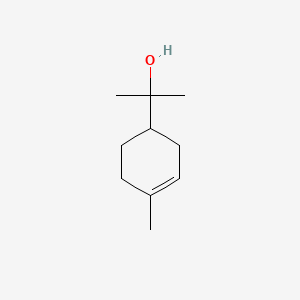 |
0.532 | D03VFL | 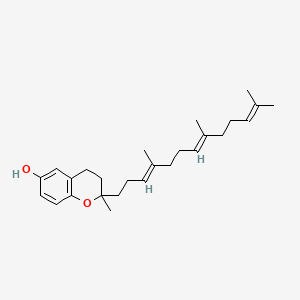 |
0.210 | ||
| ENC001981 | 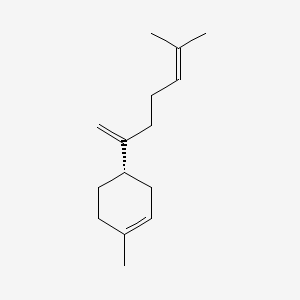 |
0.527 | D07QKN | 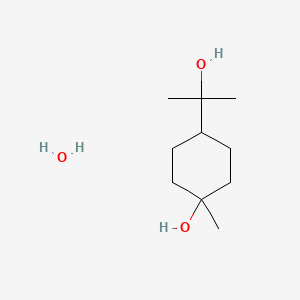 |
0.210 | ||
| ENC001641 | 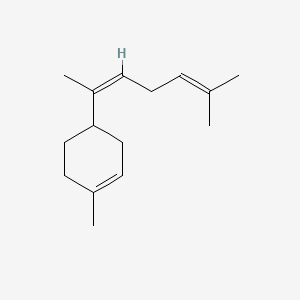 |
0.474 | D0S7WX | 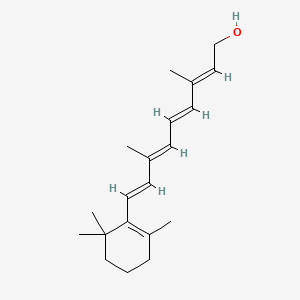 |
0.205 | ||
| ENC000952 | 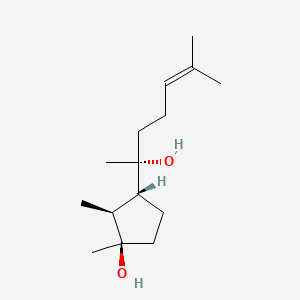 |
0.450 | D0O1UZ |  |
0.205 | ||
| ENC001484 |  |
0.424 | D0P1FO |  |
0.189 | ||
| ENC002339 |  |
0.403 | D0OK5I |  |
0.187 | ||
| ENC003092 |  |
0.381 | D05XQE |  |
0.184 | ||
| ENC001827 |  |
0.377 | D0X7XG | 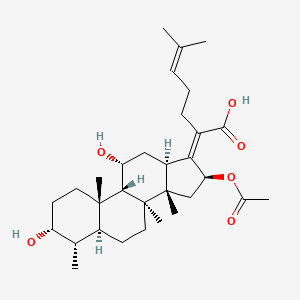 |
0.183 | ||