NPs Basic Information
|
Name |
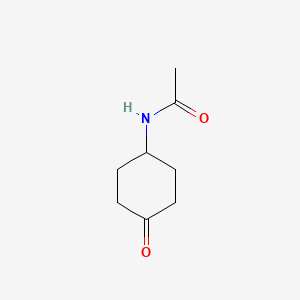
N-(4-Oxocyclohexyl)acetamide
|
| Molecular Formula | C8H13NO2 | |
| IUPAC Name* |
N-(4-oxocyclohexyl)acetamide
|
|
| SMILES |
CC(=O)NC1CCC(=O)CC1
|
|
| InChI |
InChI=1S/C8H13NO2/c1-6(10)9-7-2-4-8(11)5-3-7/h7H,2-5H2,1H3,(H,9,10)
|
|
| InChIKey |
WZEMYWNHKFIVKE-UHFFFAOYSA-N
|
|
| Synonyms |
N-(4-Oxocyclohexyl)acetamide; 27514-08-5; 4-Acetamidocyclohexanone; 4-acetamido-cyclohexanone; 4-(Acetylamino)cyclohexanone; Acetamide, N-(4-oxocyclohexyl)-; N-(4-oxocyclohexyl) acetamide; 4-n-acetyl-amino-cyclohexanone; MFCD03703462; 4-n-acetylamino cyclohexanone; n-(4-oxo-cyclohexyl)-acetamide; 4-Aminocyclohexanone, N-acetyl-; n-acetylaminocyclohexanone; 4-acetamido cyclohexanone; 4-acetylamino-cyclohexanone; EC 608-106-4; N-(4-Oxocyclohexyl)acetamde; SCHEMBL658632; 4-Acetamidocyclohexanone, 97%; N-(4-Oxocyclohexyl)acetamide #; DTXSID40337089; CS-M1862; ZINC2510356; BBL034880; STL426087; AKOS001372662; AC-5742; GS-3222; SB17822; SY020514; DB-007715; AM20070523; FT-0602731; EN300-27282; 514A085; J-523073; Z235341209; 4-N-acetyl-aMino-cyclohexanone;N-(4-Oxocyclohexyl)acetaMide;4-acetaMino cyclohexanone
|
|
| CAS | 27514-08-5 | |
| PubChem CID | 538565 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 155.19 | ALogp: | -0.2 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 46.2 | Aromatic Rings: | 1 |
| Heavy Atoms: | 11 | QED Weighted: | 0.615 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.517 | MDCK Permeability: | 0.00002260 |
| Pgp-inhibitor: | 0.016 | Pgp-substrate: | 0.643 |
| Human Intestinal Absorption (HIA): | 0.012 | 20% Bioavailability (F20%): | 0.024 |
| 30% Bioavailability (F30%): | 0.014 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.984 | Plasma Protein Binding (PPB): | 19.58% |
| Volume Distribution (VD): | 0.699 | Fu: | 85.75% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.015 | CYP1A2-substrate: | 0.129 |
| CYP2C19-inhibitor: | 0.032 | CYP2C19-substrate: | 0.129 |
| CYP2C9-inhibitor: | 0.013 | CYP2C9-substrate: | 0.343 |
| CYP2D6-inhibitor: | 0.007 | CYP2D6-substrate: | 0.255 |
| CYP3A4-inhibitor: | 0.007 | CYP3A4-substrate: | 0.168 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.988 | Half-life (T1/2): | 0.827 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.016 | Human Hepatotoxicity (H-HT): | 0.3 |
| Drug-inuced Liver Injury (DILI): | 0.067 | AMES Toxicity: | 0.364 |
| Rat Oral Acute Toxicity: | 0.013 | Maximum Recommended Daily Dose: | 0.247 |
| Skin Sensitization: | 0.129 | Carcinogencity: | 0.028 |
| Eye Corrosion: | 0.004 | Eye Irritation: | 0.087 |
| Respiratory Toxicity: | 0.027 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC004122 | 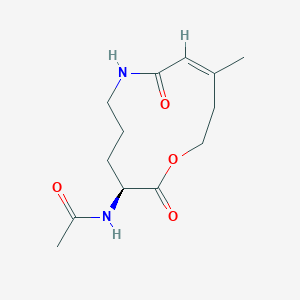 |
0.302 | D04JPJ | 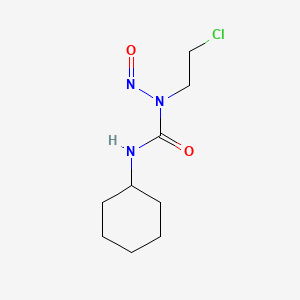 |
0.263 | ||
| ENC000567 |  |
0.255 | D07WFK | 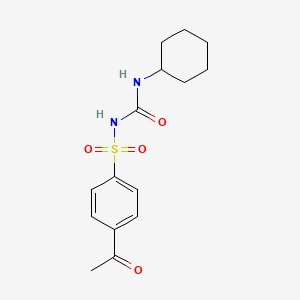 |
0.247 | ||
| ENC001082 | 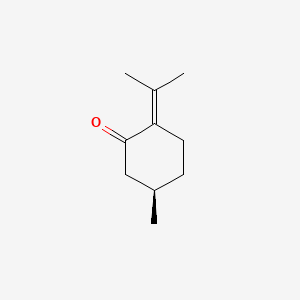 |
0.255 | D0Z8SF | 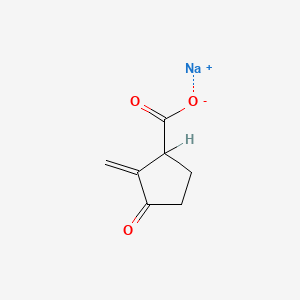 |
0.239 | ||
| ENC001339 | 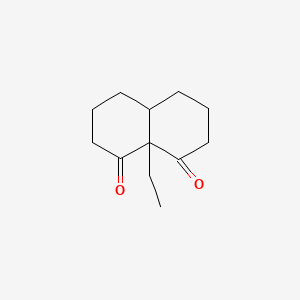 |
0.255 | D0R7WU | 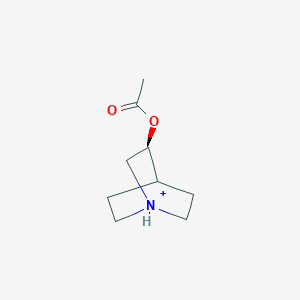 |
0.231 | ||
| ENC000828 | 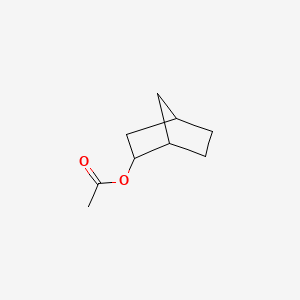 |
0.245 | D07BSQ | 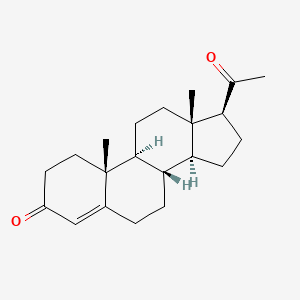 |
0.221 | ||
| ENC001302 | 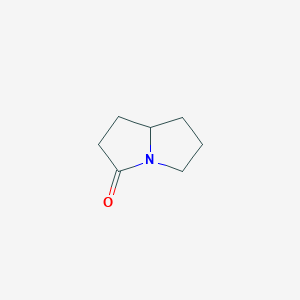 |
0.244 | D0E1XL | 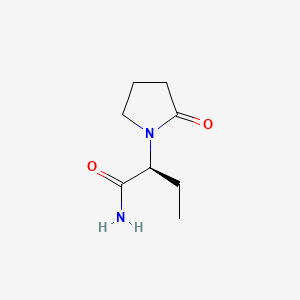 |
0.216 | ||
| ENC001191 |  |
0.241 | D0Q4YK |  |
0.213 | ||
| ENC003574 |  |
0.239 | D06FPQ |  |
0.209 | ||
| ENC000456 | 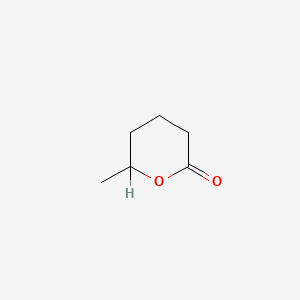 |
0.238 | D0R9BG | 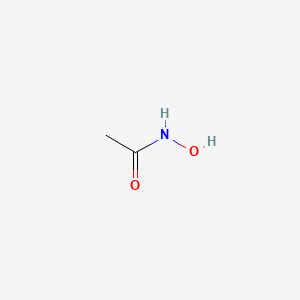 |
0.206 | ||
| ENC001256 | 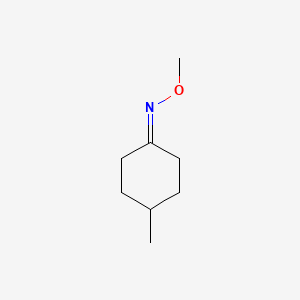 |
0.234 | D0F1UL | 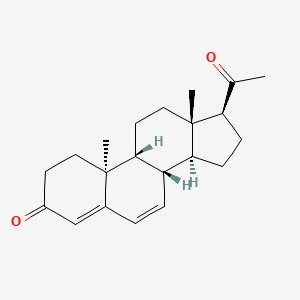 |
0.205 | ||