NPs Basic Information
|
Name |
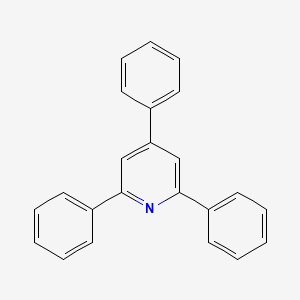
2,4,6-Triphenylpyridine
|
| Molecular Formula | C23H17N | |
| IUPAC Name* |
2,4,6-triphenylpyridine
|
|
| SMILES |
C1=CC=C(C=C1)C2=CC(=NC(=C2)C3=CC=CC=C3)C4=CC=CC=C4
|
|
| InChI |
InChI=1S/C23H17N/c1-4-10-18(11-5-1)21-16-22(19-12-6-2-7-13-19)24-23(17-21)20-14-8-3-9-15-20/h1-17H
|
|
| InChIKey |
FRZHWQQBYDFNTH-UHFFFAOYSA-N
|
|
| Synonyms |
2,4,6-Triphenylpyridine; 580-35-8; Pyridine, 2,4,6-triphenyl-; 2,4,6-Triphenyl-pyridin; 2,4,6-Triphenyl-pyridine; 2,6-Triphenylpyridine; NSC2245; Pyridine,4,6-triphenyl-; 2,4,6,-triphenylpyridine; CHEMBL27882; SCHEMBL960513; YSCH0044; DTXSID30206730; BCP15453; NSC 2245; NSC-2245; ZINC1541473; MFCD00014630; STK391423; AKOS001031352; CS-W012081; NCGC00331413-01; UPCMLD0ENAT0505-1329:001; DB-019855; FT-0609919; T3394; T72658; AB01099975-04; 2.4.6-TRICHLORO-1.3.5-TRIPHENYLBORAZINE; A831709
|
|
| CAS | 580-35-8 | |
| PubChem CID | 136370 | |
| ChEMBL ID | CHEMBL27882 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 307.4 | ALogp: | 5.8 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 12.9 | Aromatic Rings: | 4 |
| Heavy Atoms: | 24 | QED Weighted: | 0.444 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.747 | MDCK Permeability: | 0.00001470 |
| Pgp-inhibitor: | 0.896 | Pgp-substrate: | 0.97 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.997 |
| 30% Bioavailability (F30%): | 0.276 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.247 | Plasma Protein Binding (PPB): | 101.23% |
| Volume Distribution (VD): | 0.78 | Fu: | 0.74% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.979 | CYP1A2-substrate: | 0.133 |
| CYP2C19-inhibitor: | 0.607 | CYP2C19-substrate: | 0.047 |
| CYP2C9-inhibitor: | 0.291 | CYP2C9-substrate: | 0.594 |
| CYP2D6-inhibitor: | 0 | CYP2D6-substrate: | 0.088 |
| CYP3A4-inhibitor: | 0.044 | CYP3A4-substrate: | 0.17 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.948 | Half-life (T1/2): | 0.066 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.203 | Human Hepatotoxicity (H-HT): | 0.051 |
| Drug-inuced Liver Injury (DILI): | 0.531 | AMES Toxicity: | 0.552 |
| Rat Oral Acute Toxicity: | 0.403 | Maximum Recommended Daily Dose: | 0.269 |
| Skin Sensitization: | 0.651 | Carcinogencity: | 0.706 |
| Eye Corrosion: | 0.01 | Eye Irritation: | 0.959 |
| Respiratory Toxicity: | 0.034 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001050 |  |
0.420 | D0AA2D | 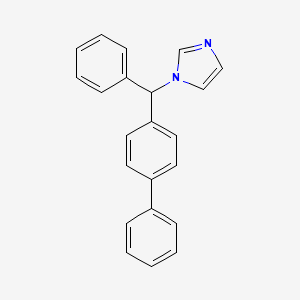 |
0.455 | ||
| ENC001402 | 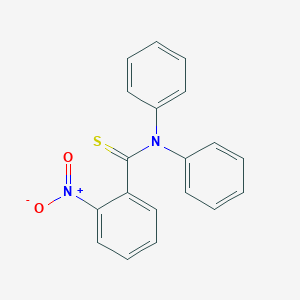 |
0.396 | D09GOS | 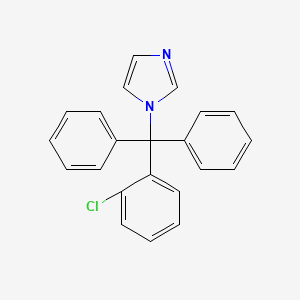 |
0.431 | ||
| ENC000321 | 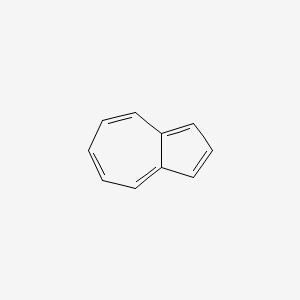 |
0.378 | D0M9DC | 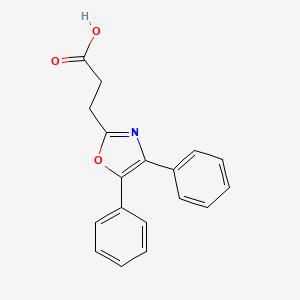 |
0.402 | ||
| ENC000732 | 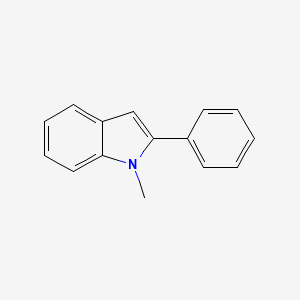 |
0.368 | D0U3ED | 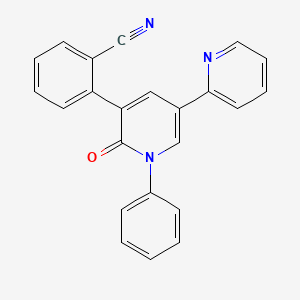 |
0.398 | ||
| ENC001018 |  |
0.364 | D03DEI |  |
0.393 | ||
| ENC003032 |  |
0.354 | D0Q3YO |  |
0.368 | ||
| ENC004519 | 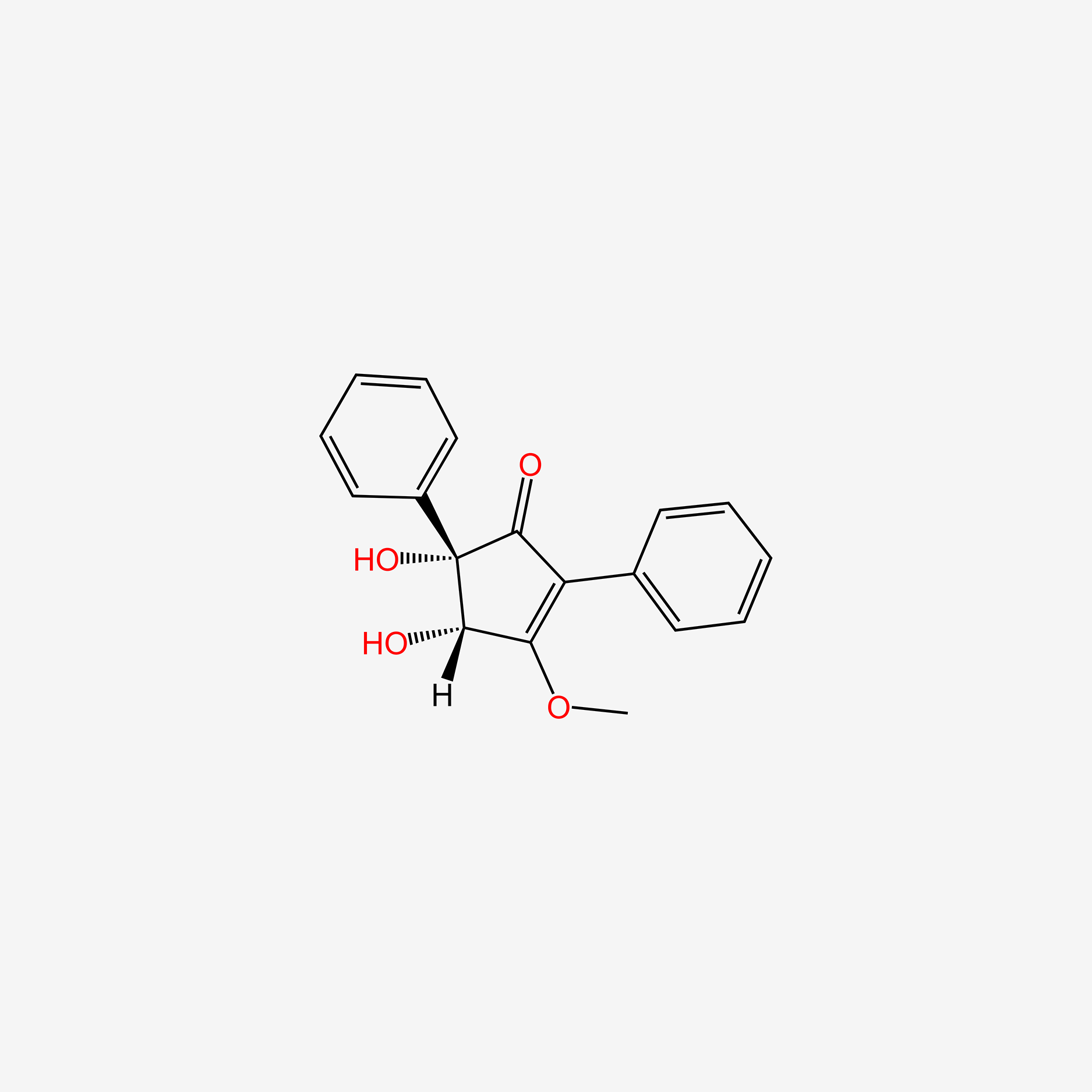 |
0.340 | D0VU2X | 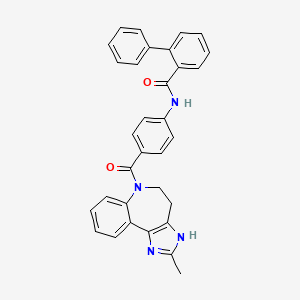 |
0.356 | ||
| ENC004518 | 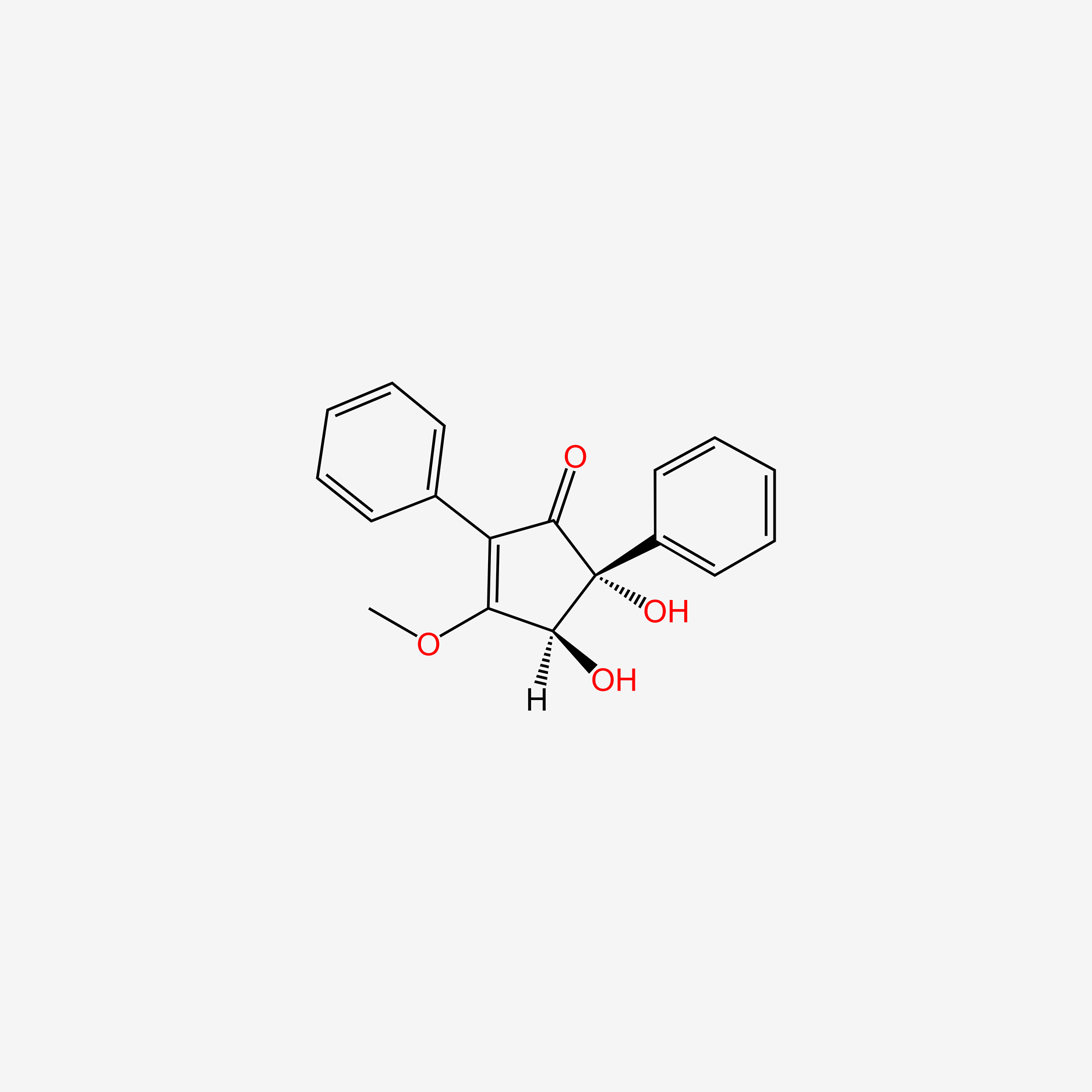 |
0.340 | D01VMO | 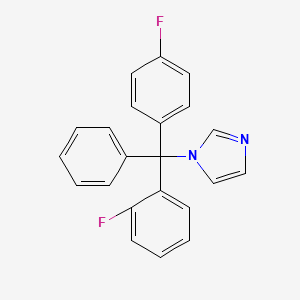 |
0.345 | ||
| ENC004517 | 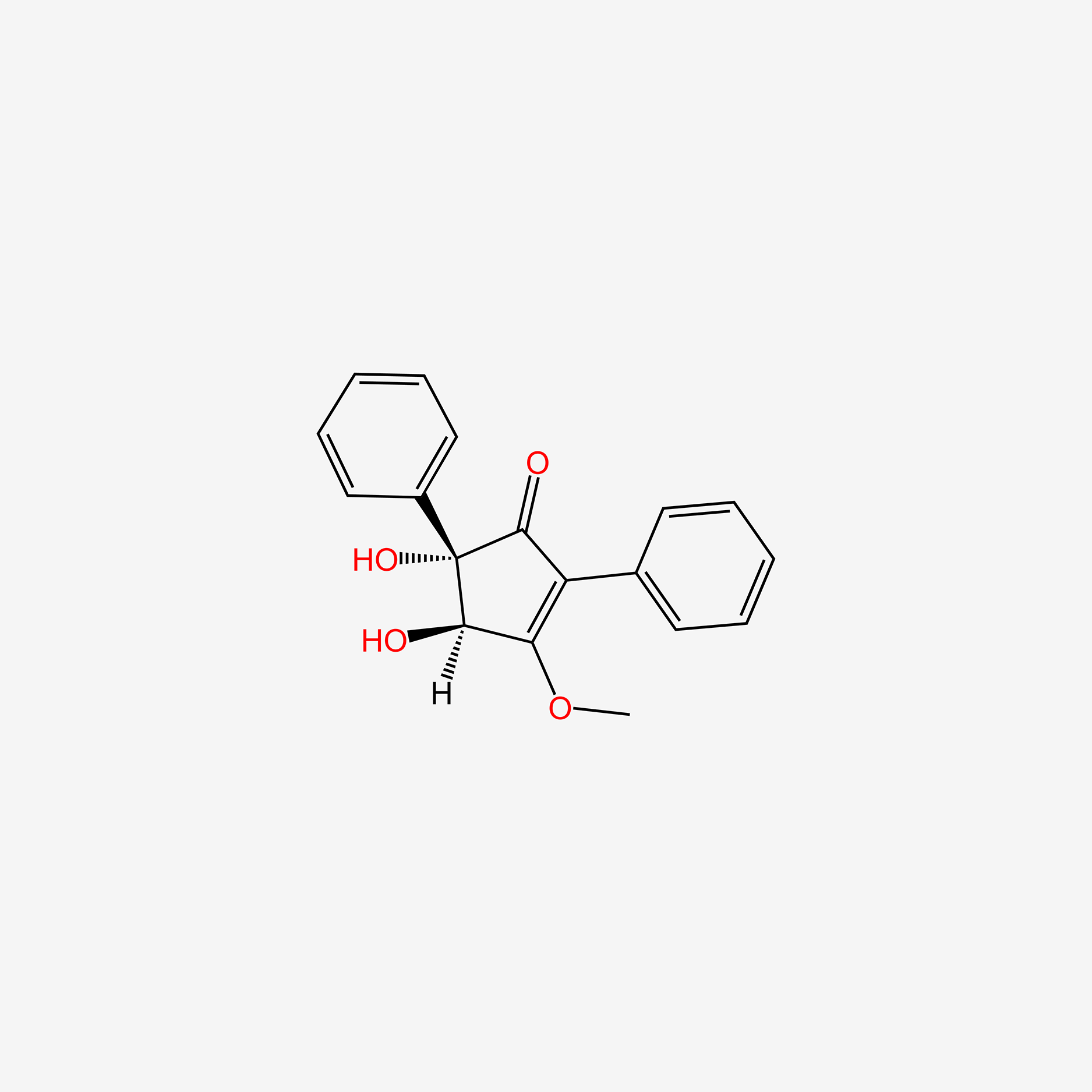 |
0.340 | D00SLY | 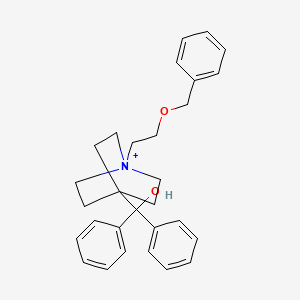 |
0.336 | ||
| ENC001752 | 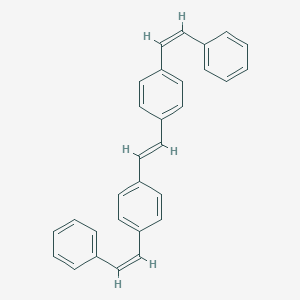 |
0.339 | D09ZOQ | 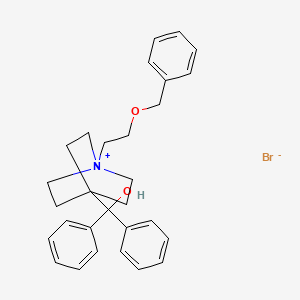 |
0.333 | ||